Summary
This strain contains 53 protein-level mutations:
- 27 mutations in Spike protein: V3G, T19I, G142D, V213G, G339D, S371F, S373P, 375-376 ST->FA, D405N, R408S, K417N, N440K, L452R, 477-478 ST->NK, E484A, F486V, Q498R, N501Y, Y505H, D614G, H655Y, N679K, P681H, N764K, D796Y, Q954H, N969K
- 5 mutations in N protein: P13L, ERS 31-33 deletion, P151S, 203-204 RG->KR, S413R
- 2 mutations in M protein: Q19E, A63T
- 1 mutation in E protein: T9I
- 1 mutation in NS3 protein: T223I
- 2 mutations in NS9b protein: P10S, ENA 27-29 deletion
- 1 mutation in NS9c protein: G50N
- 2 mutations in NSP1 protein: S135R, KSF 141-143 deletion
- 2 mutations in NSP3 protein: T24I, G489S
- 3 mutations in NSP4 protein: L264F, T327I, T492I
- 1 mutation in NSP5 protein: P132H
- 1 mutation in NSP6 protein: SGF 106-108 deletion
- 1 mutation in NSP12 protein: P323L
- 1 mutation in NSP13 protein: R392C
- 2 mutations in NSP14 protein: I42V, D301G
- 1 mutation in NSP15 protein: T112I
We identified all possible linear viral peptides affected by these mutations. Whenever it was possible, we matched the reference peptide with the mutated one. For example, D -> L mutation transformed SDNGPQNQR to SLNGPQNQR. Cases when it was not meaningful included deletions and insertions at the flanks of the peptide, e.g., HV deletion in NVTWFHAIHV peptide.
Then, we predicted binding affinities between the selected peptides and frequent HLA alleles. Predictions were made with NetMHCpan-4.1 and NetMHCIIpan-4.0. The binding affinities were classifies into three groups:
- Tight binding (IC50 affinity ≤ 50 nM)
- Moderate binding (50 nM < IC50 affinity ≤ 500 nM)
- Weak/no binding (IC50 affinity > 500 nM)
Here we report HLA-peptide interactions whose affinity was altered by at least two folds. Note that mutations with empty set of altered interactions are not showed.
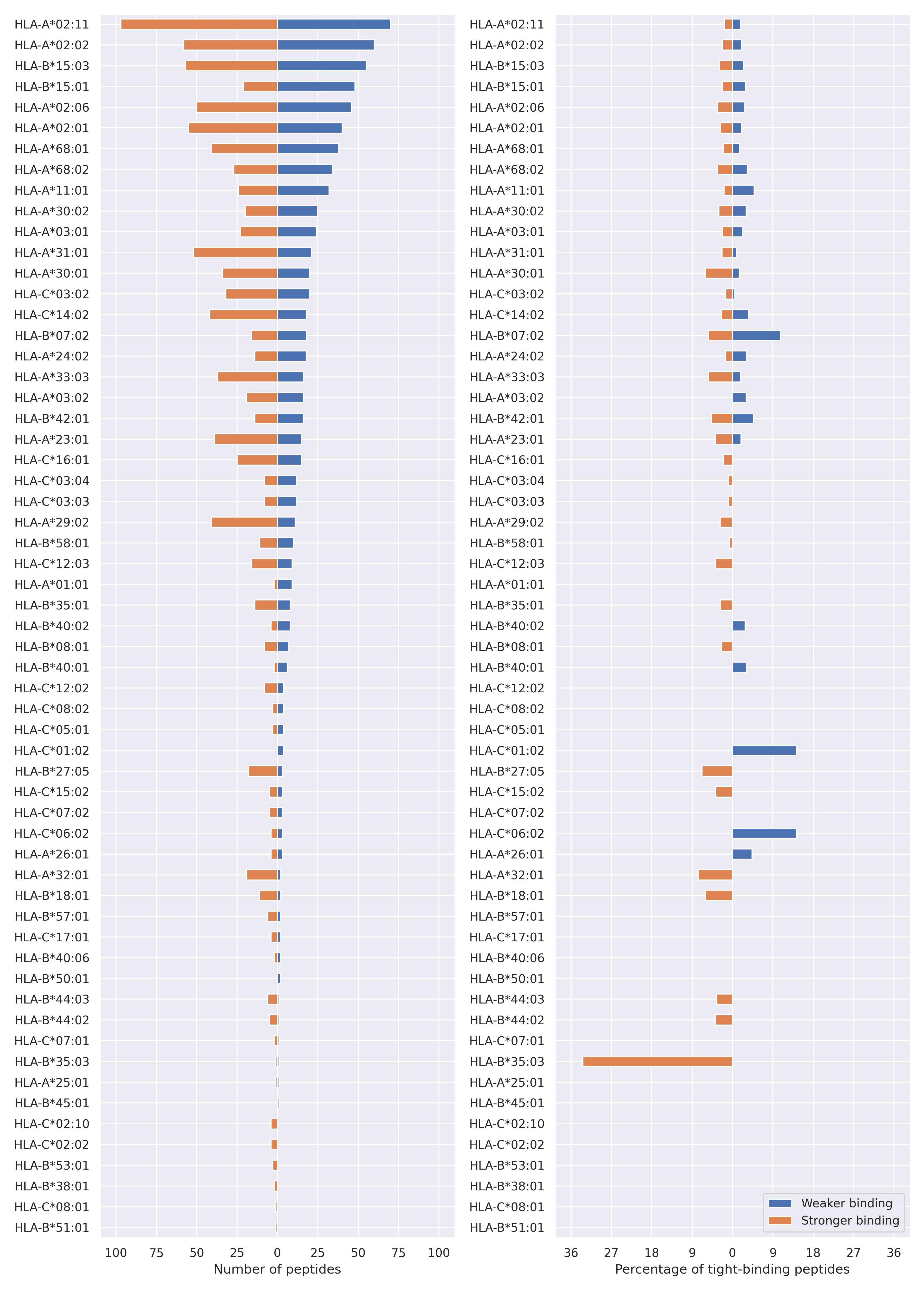
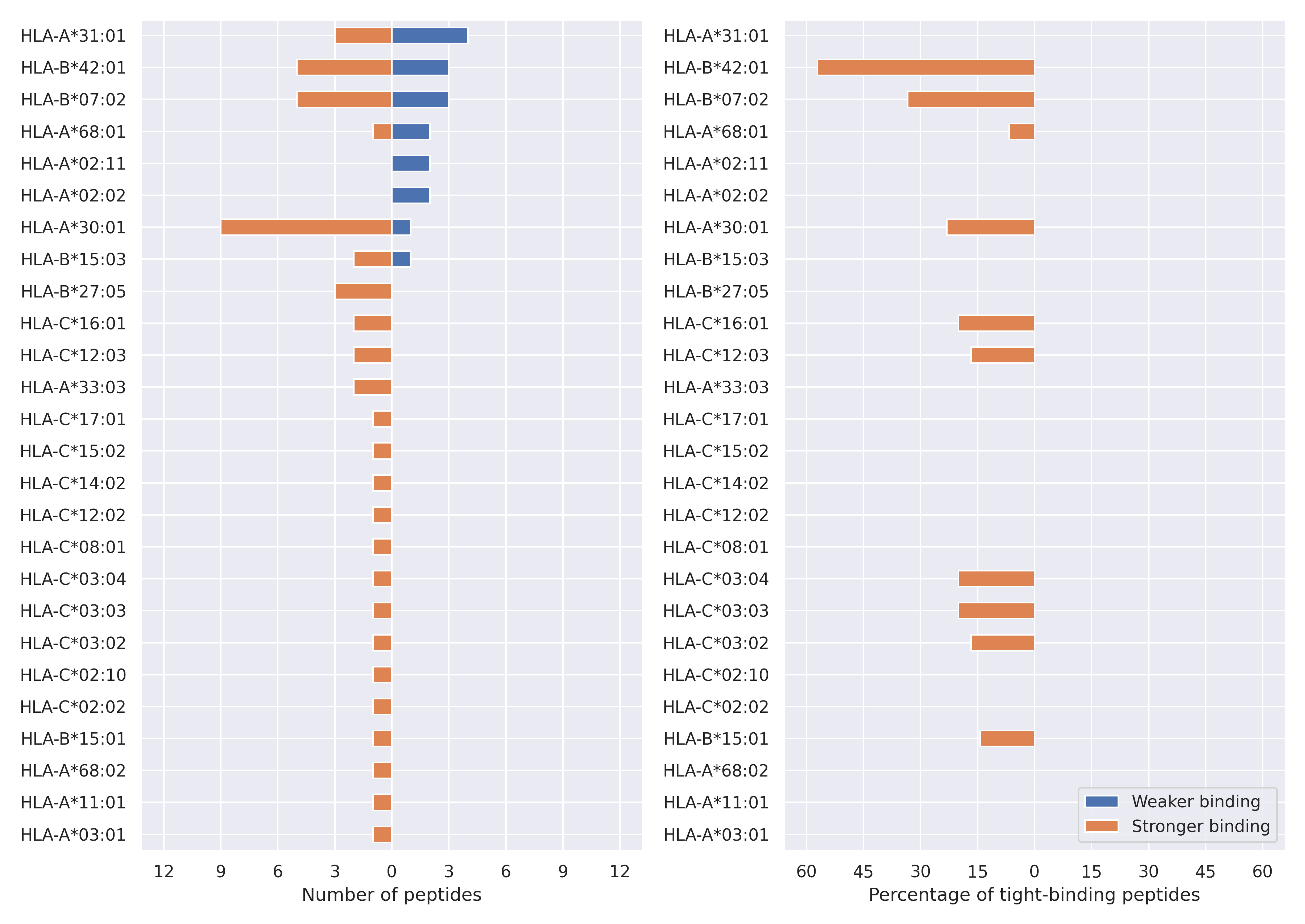
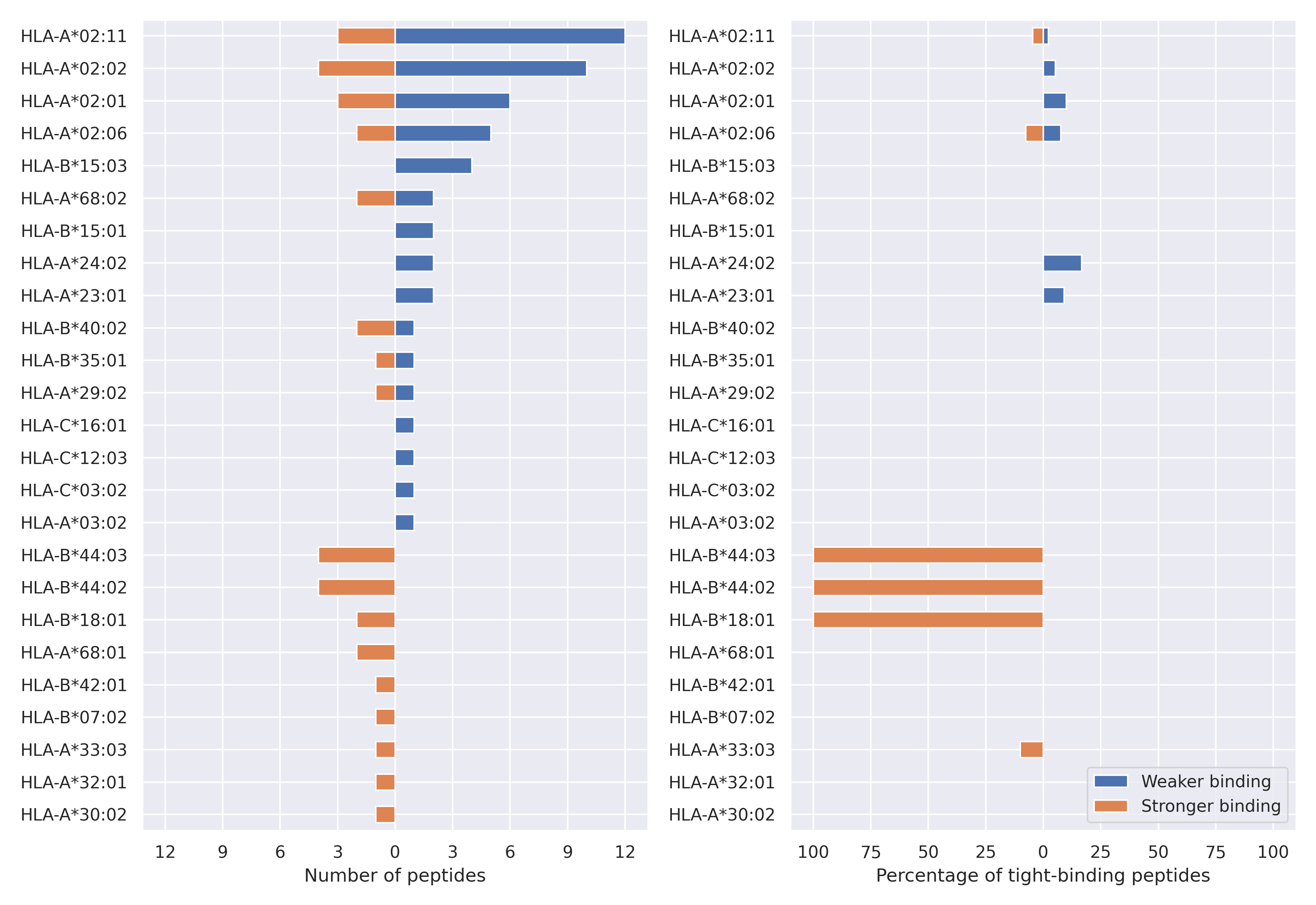
The total number of HLA-peptide interactions affected by the mutations
For each allele we calculated the number of peptides with increased and decreased affinitiy (two or more times). The absolute number of such peptides is showed on the left side of the figure (blue bars — weaker binding, orange bars — stronger).
While absolute numbers are of interest, they do not reflect the initial number of tighly binding peptides for the specific allele. Roughly speaking, if an allele had high number of tight binders, then vanished affinity of several peptides will not affect much the integral ability of peptide presentation. To the contrary, low number of vanished peptides could be important if the particular allele initially had narrow epitope repertoire. To account for this effect, we normalized the absolute numbers of new/vanished tight binders (IC50 affinity ≤ 50 nM) by the total number of tight binders for each allele. To avoid possible divisions by zero, we used + 1 regularization term in all denominators. For example, if some allele had no tight binders in the reference genome, and three new tight binders appeared as a result of the mutation, the relative increase will be 300%. The results are showed in the right part of the figure. Protein-specific plots are listed below.
Spike protein

V3G
Reference MFVFLVLLPLVSSQCVNLTTRTQLPPAY
|| |||||||||||||||||||||||||
Mutated MFGFLVLLPLVSSQCVNLITRTQLPPAY
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-A*02:11 | FVFLVLLPL | FGFLVLLPL | 17 | 584 |
| HLA-A*02:11 | FVFLVLLPLV | FGFLVLLPLV | 20 | 335 |
| HLA-A*02:01 | FVFLVLLPL | FGFLVLLPL | 125 | 1770 |
| HLA-A*02:01 | FVFLVLLPLV | FGFLVLLPLV | 62 | 835 |
| HLA-A*68:02 | FVFLVLLPL | FGFLVLLPL | 212 | 2359 |
| HLA-A*68:02 | FVFLVLLPLV | FGFLVLLPLV | 128 | 1166 |
| HLA-A*02:02 | FVFLVLLPL | FGFLVLLPL | 132 | 1193 |
| HLA-A*02:11 | MFVFLVLL | MFGFLVLL | 4346 | 496 |
| HLA-A*68:02 | MFVFLVLLPL | MFGFLVLLPL | 354 | 2873 |
| HLA-A*02:02 | FVFLVLLPLV | FGFLVLLPLV | 104 | 826 |
| HLA-A*02:06 | FVFLVLLPLV | FGFLVLLPLV | 26 | 203 |
| HLA-A*02:11 | MFVFLVLLPL | MFGFLVLLPL | 269 | 1834 |
| HLA-A*02:06 | FVFLVLLPL | FGFLVLLPL | 67 | 434 |
| HLA-A*02:06 | MFVFLVLLPL | MFGFLVLLPL | 336 | 1502 |
| HLA-A*02:02 | MFVFLVLLPL | MFGFLVLLPL | 395 | 1255 |
| HLA-C*03:02 | FVFLVLLPL | FGFLVLLPL | 79 | 214 |
| HLA-C*03:03 | FVFLVLLPL | FGFLVLLPL | 58 | 140 |
| HLA-C*03:04 | FVFLVLLPL | FGFLVLLPL | 58 | 140 |
T19I
Reference MFVFLVLLPLVSSQCVNLTTRTQLPPAYTNSFTRGVYYPDKVFR
|||||||||||||||||| |||||||||||||||||||||||||
Mutated MFGFLVLLPLVSSQCVNLITRTQLPPAYTNSFTRGVYYPDKVFR
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-B*58:01 | VSSQCVNLT | VSSQCVNLI | 3986 | 159 |
| HLA-C*15:02 | VSSQCVNLT | VSSQCVNLI | 10208 | 479 |
| HLA-A*02:02 | LVSSQCVNLT | LVSSQCVNLI | 3376 | 271 |
| HLA-A*02:11 | VLLPLVSSQCVNLT | VLLPLVSSQCVNLI | 4460 | 394 |
| HLA-A*02:11 | NLTTRTQLPPA | NLITRTQLPPA | 4965 | 468 |
| HLA-B*08:01 | NLTTRTQL | NLITRTQL | 531 | 100 |
| HLA-B*15:03 | TTRTQLPPAY | ITRTQLPPAY | 888 | 426 |
G142D
Reference LLIVNNATNVVIKVCEFQFCNDPFLGVYYHKNNKSWMESEFRVYSSANNCT
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated LLIVNNATNVVIKVCEFQFCNDPFLDVYYHKNNKSWMESEFRVYSSANNCT
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-A*03:01 | GVYYHKNNK | DVYYHKNNK | 27 | 2841 |
| HLA-A*30:01 | GVYYHKNNK | DVYYHKNNK | 51 | 3035 |
| HLA-A*11:01 | GVYYHKNNK | DVYYHKNNK | 51 | 1768 |
| HLA-A*03:02 | GVYYHKNNK | DVYYHKNNK | 119 | 3887 |
| HLA-A*31:01 | GVYYHKNNK | DVYYHKNNK | 375 | 5203 |
| HLA-A*68:01 | GVYYHKNNK | DVYYHKNNK | 363 | 28 |
| HLA-A*68:02 | FCNDPFLGV | FCNDPFLDV | 281 | 3555 |
| HLA-A*33:03 | GVYYHKNNK | DVYYHKNNK | 3253 | 287 |
| HLA-A*02:06 | FCNDPFLGV | FCNDPFLDV | 116 | 1132 |
| HLA-A*02:11 | FCNDPFLGV | FCNDPFLDV | 87 | 751 |
| HLA-A*02:06 | EFQFCNDPFLGV | EFQFCNDPFLDV | 87 | 554 |
| HLA-A*30:02 | CNDPFLGVY | CNDPFLDVY | 310 | 1959 |
| HLA-A*02:02 | FCNDPFLGV | FCNDPFLDV | 365 | 2187 |
| HLA-A*02:06 | FQFCNDPFLGVY | FQFCNDPFLDVY | 135 | 784 |
| HLA-A*30:02 | FCNDPFLGVY | FCNDPFLDVY | 408 | 1738 |
| HLA-A*02:11 | EFQFCNDPFLGV | EFQFCNDPFLDV | 334 | 1214 |
| HLA-A*02:02 | EFQFCNDPFLGV | EFQFCNDPFLDV | 477 | 1725 |
| HLA-A*02:11 | FQFCNDPFLGVY | FQFCNDPFLDVY | 488 | 1737 |
| HLA-A*02:06 | FQFCNDPFLGV | FQFCNDPFLDV | 7 | 21 |
| HLA-A*02:11 | FQFCNDPFLGV | FQFCNDPFLDV | 17 | 49 |
| HLA-A*02:01 | FQFCNDPFLGV | FQFCNDPFLDV | 39 | 111 |
| HLA-A*30:02 | FQFCNDPFLGVY | FQFCNDPFLDVY | 336 | 923 |
| HLA-A*02:02 | FQFCNDPFLGV | FQFCNDPFLDV | 45 | 122 |
| HLA-B*15:01 | FQFCNDPFLGVY | FQFCNDPFLDVY | 28 | 56 |
V213G
Reference NLREFVFKNIDGYFKIYSKHTPINLVRDLPQGFSALEPLVDLPIGINITRF
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated NLREFVFKNIDGYFKIYSKHTPINLGRDLPQGFSALEPLVDLPIGINITRF
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-A*68:02 | YSKHTPINLV | YSKHTPINLG | 199 | 16298 |
| HLA-A*02:11 | KIYSKHTPINLV | KIYSKHTPINLG | 104 | 6984 |
| HLA-B*15:01 | LVRDLPQGF | LGRDLPQGF | 160 | 2486 |
| HLA-C*01:02 | VRDLPQGFSAL | GRDLPQGFSAL | 431 | 3580 |
| HLA-C*03:02 | LVRDLPQGF | LGRDLPQGF | 259 | 1765 |
| HLA-B*15:03 | LVRDLPQGF | LGRDLPQGF | 254 | 793 |
| HLA-B*07:02 | HTPINLVRDL | HTPINLGRDL | 892 | 311 |
| HLA-B*27:05 | VRDLPQGFSA | GRDLPQGFSA | 1057 | 375 |
| HLA-A*68:01 | YSKHTPINLVR | YSKHTPINLGR | 362 | 130 |
| HLA-B*07:02 | TPINLVRDL | TPINLGRDL | 213 | 81 |
| HLA-A*68:01 | HTPINLVR | HTPINLGR | 480 | 202 |
| HLA-A*33:03 | YSKHTPINLVR | YSKHTPINLGR | 762 | 337 |
G339D
Reference QTSNFRVQPTESIVRFPNITNLCPFGEVFNATRFASVYAWNRKRISNCVAD
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated QTSNFRVQPTESIVRFPNITNLCPFDEVFNATRFASVYAWNRKRISNCVAD
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-B*40:01 | GEVFNATRF | DEVFNATRF | 164 | 7112 |
| HLA-B*18:01 | GEVFNATRFASVY | DEVFNATRFASVY | 2165 | 64 |
| HLA-B*18:01 | GEVFNATRF | DEVFNATRF | 971 | 32 |
| HLA-B*50:01 | GEVFNATRFA | DEVFNATRFA | 147 | 4233 |
| HLA-B*40:02 | GEVFNATRF | DEVFNATRF | 283 | 7625 |
| HLA-B*40:06 | GEVFNATRFA | DEVFNATRFA | 201 | 5089 |
| HLA-B*15:03 | GEVFNATRF | DEVFNATRF | 359 | 4698 |
| HLA-A*68:01 | FGEVFNATR | FDEVFNATR | 338 | 2409 |
| HLA-B*45:01 | GEVFNATRFA | DEVFNATRFA | 116 | 688 |
| HLA-B*44:02 | GEVFNATRF | DEVFNATRF | 158 | 889 |
| HLA-B*44:03 | GEVFNATRF | DEVFNATRF | 136 | 673 |
| HLA-B*15:01 | TNLCPFGEVF | TNLCPFDEVF | 368 | 1059 |
| HLA-B*15:03 | TNLCPFGEVF | TNLCPFDEVF | 442 | 1176 |
| HLA-B*15:03 | NLCPFGEVF | NLCPFDEVF | 130 | 341 |
| HLA-A*68:02 | GEVFNATRFA | DEVFNATRFA | 879 | 342 |
| HLA-B*15:01 | NLCPFGEVF | NLCPFDEVF | 315 | 773 |
S371F
Reference RFASVYAWNRKRISNCVADYSVLYNSASFSTFKCYGVSPTKLNDLCFTNVY
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated RFASVYAWNRKRISNCVADYSVLYNFAPFFAFKCYGVSPTKLNDLCFTNVY
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-A*32:01 | VLYNSASFS | VLYNFAPFF | 22186 | 33 |
| HLA-A*23:01 | VLYNSASFS | VLYNFAPFF | 32767 | 84 |
| HLA-B*15:03 | VLYNSASFS | VLYNFAPFF | 3280 | 11 |
| HLA-A*29:02 | VLYNSASFS | VLYNFAPFF | 10439 | 66 |
| HLA-A*23:01 | LYNSASFS | LYNFAPFF | 22508 | 183 |
| HLA-C*05:01 | VADYSVLYNS | VADYSVLYNF | 11802 | 122 |
| HLA-A*24:02 | LYNSASFS | LYNFAPFF | 25164 | 266 |
| HLA-A*32:01 | SVLYNSASFS | SVLYNFAPFF | 14340 | 178 |
| HLA-A*24:02 | VLYNSASFS | VLYNFAPFF | 32767 | 429 |
| HLA-A*29:02 | SVLYNSASFS | SVLYNFAPFF | 16185 | 256 |
| HLA-B*15:01 | VLYNSASFS | VLYNFAPFF | 8189 | 139 |
| HLA-C*03:02 | VLYNSASFS | VLYNFAPFF | 13004 | 222 |
| HLA-C*03:02 | VADYSVLYNS | VADYSVLYNF | 20606 | 367 |
| HLA-A*23:01 | SVLYNSASFS | SVLYNFAPFF | 24524 | 482 |
| HLA-C*14:02 | LYNSASFS | LYNFAPFF | 5693 | 117 |
| HLA-B*58:01 | VADYSVLYNS | VADYSVLYNF | 6124 | 129 |
| HLA-B*15:03 | SVLYNSASFS | SVLYNFAPFF | 2850 | 70 |
| HLA-A*01:01 | NSASFSTFKCY | NFAPFFAFKCY | 328 | 13051 |
| HLA-A*23:01 | DYSVLYNSASFS | DYSVLYNFAPFF | 14260 | 374 |
| HLA-C*16:01 | NSASFSTF | NFAPFFAF | 243 | 8562 |
| HLA-C*08:02 | VADYSVLYNS | VADYSVLYNF | 16435 | 470 |
| HLA-A*02:11 | SVLYNSASFST | SVLYNFAPFFA | 3830 | 120 |
| HLA-C*14:02 | VLYNSASFS | VLYNFAPFF | 11085 | 354 |
| HLA-A*02:01 | VLYNSASFST | VLYNFAPFFA | 243 | 8 |
| HLA-B*57:01 | VADYSVLYNS | VADYSVLYNF | 11273 | 384 |
| HLA-A*23:01 | NSASFSTF | NFAPFFAF | 12240 | 424 |
| HLA-A*29:02 | YNSASFSTF | YNFAPFFAF | 1671 | 60 |
| HLA-B*58:01 | YSVLYNSASFS | YSVLYNFAPFF | 6920 | 296 |
| HLA-A*68:02 | NSASFSTFKCYGV | NFAPFFAFKCYGV | 48 | 1043 |
| HLA-C*14:02 | NSASFSTF | NFAPFFAF | 2544 | 120 |
| HLA-B*15:01 | SVLYNSASFS | SVLYNFAPFF | 2838 | 139 |
| HLA-A*02:02 | VLYNSASFST | VLYNFAPFFA | 202 | 10 |
| HLA-C*03:02 | SVLYNSASFS | SVLYNFAPFF | 8888 | 443 |
| HLA-A*02:06 | VLYNSASFST | VLYNFAPFFA | 710 | 36 |
| HLA-B*15:03 | YSVLYNSASFS | YSVLYNFAPFF | 3170 | 164 |
| HLA-A*02:11 | VLYNSASFST | VLYNFAPFFA | 77 | 4 |
| HLA-A*02:01 | SVLYNSASFST | SVLYNFAPFFA | 5674 | 301 |
| HLA-A*11:01 | LYNSASFSTFK | LYNFAPFFAFK | 43 | 797 |
| HLA-B*40:02 | ADYSVLYNS | ADYSVLYNF | 4645 | 259 |
| HLA-A*30:02 | SASFSTFKCY | FAPFFAFKCY | 71 | 1241 |
| HLA-A*02:11 | YSVLYNSASFST | YSVLYNFAPFFA | 7859 | 469 |
| HLA-A*32:01 | YNSASFSTF | YNFAPFFAF | 4333 | 294 |
| HLA-A*33:03 | YNSASFSTFK | YNFAPFFAFK | 823 | 57 |
| HLA-A*02:06 | SVLYNSASFST | SVLYNFAPFFA | 4097 | 319 |
| HLA-B*18:01 | YNSASFSTF | YNFAPFFAF | 3495 | 284 |
| HLA-A*33:03 | NSASFSTFK | NFAPFFAFK | 181 | 16 |
| HLA-A*02:11 | VLYNSASFSTF | VLYNFAPFFAF | 2537 | 236 |
| HLA-A*11:01 | NSASFSTFK | NFAPFFAFK | 9 | 94 |
| HLA-A*29:02 | NSASFSTFKCY | NFAPFFAFKCY | 662 | 64 |
| HLA-A*29:02 | YNSASFSTFKCY | YNFAPFFAFKCY | 842 | 86 |
| HLA-A*02:01 | VLYNSASFSTF | VLYNFAPFFAF | 4199 | 455 |
| HLA-C*03:02 | YSVLYNSASFS | YSVLYNFAPFF | 2405 | 264 |
| HLA-A*02:06 | SVLYNSASF | SVLYNFAPF | 2112 | 234 |
| HLA-B*15:01 | VLYNSASFSTF | VLYNFAPFFAF | 50 | 420 |
| HLA-A*31:01 | YNSASFSTFK | YNFAPFFAFK | 607 | 73 |
| HLA-A*30:01 | LYNSASFST | LYNFAPFFA | 3467 | 433 |
| HLA-A*33:03 | SVLYNSASFSTFK | SVLYNFAPFFAFK | 2778 | 357 |
| HLA-B*35:01 | SASFSTFKCY | FAPFFAFKCY | 490 | 65 |
| HLA-A*31:01 | NSASFSTFK | NFAPFFAFK | 293 | 47 |
| HLA-A*29:02 | LYNSASFSTF | LYNFAPFFAF | 387 | 63 |
| HLA-A*30:02 | VLYNSASFS | VLYNFAPFF | 2919 | 476 |
| HLA-A*31:01 | SVLYNSASFSTFK | SVLYNFAPFFAFK | 857 | 148 |
| HLA-A*23:01 | YNSASFSTF | YNFAPFFAF | 352 | 63 |
| HLA-A*03:01 | LYNSASFSTFK | LYNFAPFFAFK | 295 | 1614 |
| HLA-A*23:01 | SVLYNSASF | SVLYNFAPF | 990 | 197 |
| HLA-A*02:11 | SVLYNSASFS | SVLYNFAPFF | 2012 | 435 |
| HLA-A*03:01 | NSASFSTFK | NFAPFFAFK | 113 | 494 |
| HLA-B*35:01 | YNSASFSTF | YNFAPFFAF | 760 | 180 |
| HLA-A*03:02 | LYNSASFSTFK | LYNFAPFFAFK | 202 | 831 |
| HLA-A*02:01 | VLYNSASFS | VLYNFAPFF | 1548 | 377 |
| HLA-A*02:11 | VLYNSASFS | VLYNFAPFF | 258 | 63 |
| HLA-A*01:01 | SASFSTFKCY | FAPFFAFKCY | 461 | 1849 |
| HLA-A*11:01 | SASFSTFK | FAPFFAFK | 452 | 1795 |
| HLA-A*68:01 | SASFSTFK | FAPFFAFK | 1114 | 290 |
| HLA-B*15:01 | YSVLYNSASFSTF | YSVLYNFAPFFAF | 164 | 629 |
| HLA-A*03:02 | NSASFSTFK | NFAPFFAFK | 37 | 135 |
| HLA-A*29:02 | SVLYNSASF | SVLYNFAPF | 1130 | 313 |
| HLA-A*30:02 | NSASFSTFKCY | NFAPFFAFKCY | 387 | 1395 |
| HLA-C*12:03 | SASFSTFKCY | FAPFFAFKCY | 498 | 139 |
| HLA-C*14:02 | LYNSASFSTF | LYNFAPFFAF | 20 | 69 |
| HLA-A*30:02 | VLYNSASFSTFKCY | VLYNFAPFFAFKCY | 272 | 923 |
| HLA-A*32:01 | SVLYNSASFSTF | SVLYNFAPFFAF | 1624 | 488 |
| HLA-A*31:01 | VLYNSASFSTFK | VLYNFAPFFAFK | 565 | 170 |
| HLA-A*68:01 | NSASFSTFK | NFAPFFAFK | 4 | 13 |
| HLA-A*31:01 | LYNSASFSTFK | LYNFAPFFAFK | 299 | 92 |
| HLA-A*11:01 | YNSASFSTFK | YNFAPFFAFK | 12 | 38 |
| HLA-A*24:02 | LYNSASFSTFK | LYNFAPFFAFK | 356 | 1101 |
| HLA-A*26:01 | SVLYNSASF | SVLYNFAPF | 1313 | 431 |
| HLA-A*33:03 | LYNSASFSTFK | LYNFAPFFAFK | 1158 | 383 |
| HLA-A*24:02 | VLYNSASFSTF | VLYNFAPFFAF | 186 | 551 |
| HLA-A*03:01 | VLYNSASFSTFKC | VLYNFAPFFAFKC | 312 | 920 |
| HLA-A*02:06 | SASFSTFKCYGV | FAPFFAFKCYGV | 1416 | 481 |
| HLA-C*03:02 | YSVLYNSASFSTF | YSVLYNFAPFFAF | 113 | 329 |
| HLA-A*23:01 | YSVLYNSASF | YSVLYNFAPF | 1120 | 403 |
| HLA-B*58:01 | YSVLYNSASFSTF | YSVLYNFAPFFAF | 429 | 1160 |
| HLA-A*29:02 | LYNSASFSTFKCY | LYNFAPFFAFKCY | 297 | 115 |
| HLA-C*12:02 | SASFSTFKCY | FAPFFAFKCY | 710 | 275 |
| HLA-B*15:03 | VLYNSASFSTF | VLYNFAPFFAF | 38 | 96 |
| HLA-A*68:01 | NSASFSTFKC | NFAPFFAFKC | 281 | 702 |
| HLA-A*03:01 | VLYNSASFSTFK | VLYNFAPFFAFK | 7 | 17 |
| HLA-A*03:01 | YNSASFSTFK | YNFAPFFAFK | 80 | 194 |
| HLA-A*11:01 | VLYNSASFSTFK | VLYNFAPFFAFK | 22 | 51 |
| HLA-B*15:01 | SVLYNSASFSTF | SVLYNFAPFFAF | 221 | 509 |
| HLA-B*15:01 | LYNSASFSTF | LYNFAPFFAF | 468 | 1064 |
| HLA-B*15:01 | SASFSTFKCY | FAPFFAFKCY | 256 | 561 |
| HLA-C*03:02 | SASFSTFKCY | FAPFFAFKCY | 212 | 97 |
| HLA-A*23:01 | DYSVLYNSASF | DYSVLYNFAPF | 1039 | 482 |
| HLA-A*68:01 | YSVLYNSASFSTFK | YSVLYNFAPFFAFK | 144 | 67 |
S373P
Reference ASVYAWNRKRISNCVADYSVLYNSASFSTFKCYGVSPTKLNDLCFTNVYAD
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated ASVYAWNRKRISNCVADYSVLYNFAPFFAFKCYGVSPTKLNDLCFTNVYAD
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-A*32:01 | VLYNSASFS | VLYNFAPFF | 22186 | 33 |
| HLA-A*23:01 | VLYNSASFS | VLYNFAPFF | 32767 | 84 |
| HLA-B*15:03 | VLYNSASFS | VLYNFAPFF | 3280 | 11 |
| HLA-A*29:02 | VLYNSASFS | VLYNFAPFF | 10439 | 66 |
| HLA-A*23:01 | LYNSASFS | LYNFAPFF | 22508 | 183 |
| HLA-A*24:02 | LYNSASFS | LYNFAPFF | 25164 | 266 |
| HLA-A*32:01 | SVLYNSASFS | SVLYNFAPFF | 14340 | 178 |
| HLA-A*24:02 | VLYNSASFS | VLYNFAPFF | 32767 | 429 |
| HLA-A*29:02 | SVLYNSASFS | SVLYNFAPFF | 16185 | 256 |
| HLA-A*30:02 | ASFSTFKCY | APFFAFKCY | 43 | 2647 |
| HLA-B*15:01 | VLYNSASFS | VLYNFAPFF | 8189 | 139 |
| HLA-C*03:02 | VLYNSASFS | VLYNFAPFF | 13004 | 222 |
| HLA-B*15:01 | ASFSTFKCY | APFFAFKCY | 107 | 6085 |
| HLA-A*23:01 | SVLYNSASFS | SVLYNFAPFF | 24524 | 482 |
| HLA-A*02:06 | ASFSTFKCYGV | APFFAFKCYGV | 231 | 11537 |
| HLA-C*14:02 | LYNSASFS | LYNFAPFF | 5693 | 117 |
| HLA-C*16:01 | ASFSTFKCY | APFFAFKCY | 68 | 3117 |
| HLA-C*12:03 | ASFSTFKCY | APFFAFKCY | 115 | 5066 |
| HLA-C*03:02 | ASFSTFKCY | APFFAFKCY | 64 | 2656 |
| HLA-B*15:03 | SVLYNSASFS | SVLYNFAPFF | 2850 | 70 |
| HLA-A*01:01 | NSASFSTFKCY | NFAPFFAFKCY | 328 | 13051 |
| HLA-A*23:01 | DYSVLYNSASFS | DYSVLYNFAPFF | 14260 | 374 |
| HLA-C*16:01 | NSASFSTF | NFAPFFAF | 243 | 8562 |
| HLA-A*11:01 | ASFSTFKCY | APFFAFKCY | 379 | 12606 |
| HLA-A*02:11 | SVLYNSASFST | SVLYNFAPFFA | 3830 | 120 |
| HLA-C*14:02 | VLYNSASFS | VLYNFAPFF | 11085 | 354 |
| HLA-A*02:01 | VLYNSASFST | VLYNFAPFFA | 243 | 8 |
| HLA-A*23:01 | NSASFSTF | NFAPFFAF | 12240 | 424 |
| HLA-A*29:02 | YNSASFSTF | YNFAPFFAF | 1671 | 60 |
| HLA-B*35:01 | ASFSTFKCY | APFFAFKCY | 2345 | 92 |
| HLA-C*12:02 | ASFSTFKCY | APFFAFKCY | 338 | 8222 |
| HLA-B*58:01 | YSVLYNSASFS | YSVLYNFAPFF | 6920 | 296 |
| HLA-A*68:02 | NSASFSTFKCYGV | NFAPFFAFKCYGV | 48 | 1043 |
| HLA-C*14:02 | NSASFSTF | NFAPFFAF | 2544 | 120 |
| HLA-B*15:03 | ASFSTFKCY | APFFAFKCY | 25 | 529 |
| HLA-B*15:01 | SVLYNSASFS | SVLYNFAPFF | 2838 | 139 |
| HLA-A*02:02 | VLYNSASFST | VLYNFAPFFA | 202 | 10 |
| HLA-C*03:02 | SVLYNSASFS | SVLYNFAPFF | 8888 | 443 |
| HLA-A*02:06 | VLYNSASFST | VLYNFAPFFA | 710 | 36 |
| HLA-B*15:03 | YSVLYNSASFS | YSVLYNFAPFF | 3170 | 164 |
| HLA-A*02:11 | VLYNSASFST | VLYNFAPFFA | 77 | 4 |
| HLA-A*02:01 | SVLYNSASFST | SVLYNFAPFFA | 5674 | 301 |
| HLA-A*11:01 | LYNSASFSTFK | LYNFAPFFAFK | 43 | 797 |
| HLA-A*30:02 | SASFSTFKCY | FAPFFAFKCY | 71 | 1241 |
| HLA-A*02:11 | YSVLYNSASFST | YSVLYNFAPFFA | 7859 | 469 |
| HLA-C*14:02 | SFSTFKCY | PFFAFKCY | 386 | 6449 |
| HLA-A*32:01 | YNSASFSTF | YNFAPFFAF | 4333 | 294 |
| HLA-A*33:03 | YNSASFSTFK | YNFAPFFAFK | 823 | 57 |
| HLA-A*02:06 | SVLYNSASFST | SVLYNFAPFFA | 4097 | 319 |
| HLA-B*18:01 | YNSASFSTF | YNFAPFFAF | 3495 | 284 |
| HLA-A*33:03 | NSASFSTFK | NFAPFFAFK | 181 | 16 |
| HLA-A*02:11 | VLYNSASFSTF | VLYNFAPFFAF | 2537 | 236 |
| HLA-A*11:01 | NSASFSTFK | NFAPFFAFK | 9 | 94 |
| HLA-A*29:02 | NSASFSTFKCY | NFAPFFAFKCY | 662 | 64 |
| HLA-A*29:02 | YNSASFSTFKCY | YNFAPFFAFKCY | 842 | 86 |
| HLA-A*02:01 | VLYNSASFSTF | VLYNFAPFFAF | 4199 | 455 |
| HLA-C*03:02 | YSVLYNSASFS | YSVLYNFAPFF | 2405 | 264 |
| HLA-A*29:02 | ASFSTFKCY | APFFAFKCY | 201 | 1830 |
| HLA-A*02:06 | SVLYNSASF | SVLYNFAPF | 2112 | 234 |
| HLA-B*15:01 | VLYNSASFSTF | VLYNFAPFFAF | 50 | 420 |
| HLA-A*31:01 | YNSASFSTFK | YNFAPFFAFK | 607 | 73 |
| HLA-A*30:01 | LYNSASFST | LYNFAPFFA | 3467 | 433 |
| HLA-A*33:03 | SVLYNSASFSTFK | SVLYNFAPFFAFK | 2778 | 357 |
| HLA-B*35:01 | SASFSTFKCY | FAPFFAFKCY | 490 | 65 |
| HLA-A*31:01 | NSASFSTFK | NFAPFFAFK | 293 | 47 |
| HLA-A*29:02 | LYNSASFSTF | LYNFAPFFAF | 387 | 63 |
| HLA-A*30:02 | VLYNSASFS | VLYNFAPFF | 2919 | 476 |
| HLA-A*31:01 | SVLYNSASFSTFK | SVLYNFAPFFAFK | 857 | 148 |
| HLA-A*23:01 | YNSASFSTF | YNFAPFFAF | 352 | 63 |
| HLA-A*03:01 | LYNSASFSTFK | LYNFAPFFAFK | 295 | 1614 |
| HLA-A*23:01 | SVLYNSASF | SVLYNFAPF | 990 | 197 |
| HLA-A*02:11 | SVLYNSASFS | SVLYNFAPFF | 2012 | 435 |
| HLA-A*03:01 | NSASFSTFK | NFAPFFAFK | 113 | 494 |
| HLA-B*35:01 | YNSASFSTF | YNFAPFFAF | 760 | 180 |
| HLA-A*03:02 | LYNSASFSTFK | LYNFAPFFAFK | 202 | 831 |
| HLA-A*02:01 | VLYNSASFS | VLYNFAPFF | 1548 | 377 |
| HLA-A*02:11 | VLYNSASFS | VLYNFAPFF | 258 | 63 |
| HLA-A*01:01 | SASFSTFKCY | FAPFFAFKCY | 461 | 1849 |
| HLA-A*11:01 | SASFSTFK | FAPFFAFK | 452 | 1795 |
| HLA-A*68:01 | SASFSTFK | FAPFFAFK | 1114 | 290 |
| HLA-B*15:01 | YSVLYNSASFSTF | YSVLYNFAPFFAF | 164 | 629 |
| HLA-A*03:02 | NSASFSTFK | NFAPFFAFK | 37 | 135 |
| HLA-A*29:02 | SVLYNSASF | SVLYNFAPF | 1130 | 313 |
| HLA-A*30:02 | NSASFSTFKCY | NFAPFFAFKCY | 387 | 1395 |
| HLA-C*12:03 | SASFSTFKCY | FAPFFAFKCY | 498 | 139 |
| HLA-C*14:02 | LYNSASFSTF | LYNFAPFFAF | 20 | 69 |
| HLA-A*30:02 | VLYNSASFSTFKCY | VLYNFAPFFAFKCY | 272 | 923 |
| HLA-A*32:01 | SVLYNSASFSTF | SVLYNFAPFFAF | 1624 | 488 |
| HLA-A*31:01 | VLYNSASFSTFK | VLYNFAPFFAFK | 565 | 170 |
| HLA-A*68:01 | NSASFSTFK | NFAPFFAFK | 4 | 13 |
| HLA-A*31:01 | LYNSASFSTFK | LYNFAPFFAFK | 299 | 92 |
| HLA-A*11:01 | YNSASFSTFK | YNFAPFFAFK | 12 | 38 |
| HLA-A*24:02 | LYNSASFSTFK | LYNFAPFFAFK | 356 | 1101 |
| HLA-A*26:01 | SVLYNSASF | SVLYNFAPF | 1313 | 431 |
| HLA-A*33:03 | LYNSASFSTFK | LYNFAPFFAFK | 1158 | 383 |
| HLA-A*24:02 | VLYNSASFSTF | VLYNFAPFFAF | 186 | 551 |
| HLA-A*03:01 | VLYNSASFSTFKC | VLYNFAPFFAFKC | 312 | 920 |
| HLA-A*02:06 | SASFSTFKCYGV | FAPFFAFKCYGV | 1416 | 481 |
| HLA-C*03:02 | YSVLYNSASFSTF | YSVLYNFAPFFAF | 113 | 329 |
| HLA-A*23:01 | YSVLYNSASF | YSVLYNFAPF | 1120 | 403 |
| HLA-B*58:01 | YSVLYNSASFSTF | YSVLYNFAPFFAF | 429 | 1160 |
| HLA-A*29:02 | LYNSASFSTFKCY | LYNFAPFFAFKCY | 297 | 115 |
| HLA-C*12:02 | SASFSTFKCY | FAPFFAFKCY | 710 | 275 |
| HLA-B*15:03 | VLYNSASFSTF | VLYNFAPFFAF | 38 | 96 |
| HLA-A*68:01 | NSASFSTFKC | NFAPFFAFKC | 281 | 702 |
| HLA-A*03:01 | VLYNSASFSTFK | VLYNFAPFFAFK | 7 | 17 |
| HLA-A*03:01 | YNSASFSTFK | YNFAPFFAFK | 80 | 194 |
| HLA-A*11:01 | VLYNSASFSTFK | VLYNFAPFFAFK | 22 | 51 |
| HLA-B*15:01 | SVLYNSASFSTF | SVLYNFAPFFAF | 221 | 509 |
| HLA-B*15:01 | LYNSASFSTF | LYNFAPFFAF | 468 | 1064 |
| HLA-B*15:01 | SASFSTFKCY | FAPFFAFKCY | 256 | 561 |
| HLA-C*03:02 | SASFSTFKCY | FAPFFAFKCY | 212 | 97 |
| HLA-A*23:01 | DYSVLYNSASF | DYSVLYNFAPF | 1039 | 482 |
| HLA-A*68:01 | YSVLYNSASFSTFK | YSVLYNFAPFFAFK | 144 | 67 |
375-376 ST->FA
Reference VYAWNRKRISNCVADYSVLYNSASFSTFKCYGVSPTKLNDLCFTNVYADSFV
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated VYAWNRKRISNCVADYSVLYNFAPFFAFKCYGVSPTKLNDLCFTNVYADSFV
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-A*32:01 | VLYNSASFS | VLYNFAPFF | 22186 | 33 |
| HLA-A*23:01 | VLYNSASFS | VLYNFAPFF | 32767 | 84 |
| HLA-B*15:03 | VLYNSASFS | VLYNFAPFF | 3280 | 11 |
| HLA-A*29:02 | VLYNSASFS | VLYNFAPFF | 10439 | 66 |
| HLA-A*23:01 | LYNSASFS | LYNFAPFF | 22508 | 183 |
| HLA-A*24:02 | LYNSASFS | LYNFAPFF | 25164 | 266 |
| HLA-A*32:01 | SVLYNSASFS | SVLYNFAPFF | 14340 | 178 |
| HLA-A*24:02 | VLYNSASFS | VLYNFAPFF | 32767 | 429 |
| HLA-A*29:02 | SVLYNSASFS | SVLYNFAPFF | 16185 | 256 |
| HLA-A*30:02 | ASFSTFKCY | APFFAFKCY | 43 | 2647 |
| HLA-B*15:01 | VLYNSASFS | VLYNFAPFF | 8189 | 139 |
| HLA-C*03:02 | VLYNSASFS | VLYNFAPFF | 13004 | 222 |
| HLA-B*15:01 | ASFSTFKCY | APFFAFKCY | 107 | 6085 |
| HLA-A*23:01 | SVLYNSASFS | SVLYNFAPFF | 24524 | 482 |
| HLA-A*02:06 | ASFSTFKCYGV | APFFAFKCYGV | 231 | 11537 |
| HLA-C*14:02 | LYNSASFS | LYNFAPFF | 5693 | 117 |
| HLA-C*15:02 | FSTFKCYGV | FFAFKCYGV | 431 | 20837 |
| HLA-C*16:01 | ASFSTFKCY | APFFAFKCY | 68 | 3117 |
| HLA-C*12:03 | ASFSTFKCY | APFFAFKCY | 115 | 5066 |
| HLA-C*03:02 | ASFSTFKCY | APFFAFKCY | 64 | 2656 |
| HLA-A*11:01 | STFKCYGVSPTK | FAFKCYGVSPTK | 25 | 1030 |
| HLA-B*15:03 | SVLYNSASFS | SVLYNFAPFF | 2850 | 70 |
| HLA-A*01:01 | NSASFSTFKCY | NFAPFFAFKCY | 328 | 13051 |
| HLA-A*23:01 | DYSVLYNSASFS | DYSVLYNFAPFF | 14260 | 374 |
| HLA-C*16:01 | NSASFSTF | NFAPFFAF | 243 | 8562 |
| HLA-A*11:01 | ASFSTFKCY | APFFAFKCY | 379 | 12606 |
| HLA-A*02:11 | SVLYNSASFST | SVLYNFAPFFA | 3830 | 120 |
| HLA-C*14:02 | VLYNSASFS | VLYNFAPFF | 11085 | 354 |
| HLA-A*02:01 | VLYNSASFST | VLYNFAPFFA | 243 | 8 |
| HLA-A*23:01 | NSASFSTF | NFAPFFAF | 12240 | 424 |
| HLA-A*29:02 | YNSASFSTF | YNFAPFFAF | 1671 | 60 |
| HLA-C*16:01 | FSTFKCYGV | FFAFKCYGV | 302 | 8277 |
| HLA-B*35:01 | ASFSTFKCY | APFFAFKCY | 2345 | 92 |
| HLA-C*12:02 | ASFSTFKCY | APFFAFKCY | 338 | 8222 |
| HLA-B*58:01 | YSVLYNSASFS | YSVLYNFAPFF | 6920 | 296 |
| HLA-A*03:01 | STFKCYGVSPTK | FAFKCYGVSPTK | 83 | 1880 |
| HLA-A*68:02 | NSASFSTFKCYGV | NFAPFFAFKCYGV | 48 | 1043 |
| HLA-C*14:02 | NSASFSTF | NFAPFFAF | 2544 | 120 |
| HLA-B*15:03 | ASFSTFKCY | APFFAFKCY | 25 | 529 |
| HLA-B*15:01 | SVLYNSASFS | SVLYNFAPFF | 2838 | 139 |
| HLA-A*02:02 | VLYNSASFST | VLYNFAPFFA | 202 | 10 |
| HLA-C*03:02 | SVLYNSASFS | SVLYNFAPFF | 8888 | 443 |
| HLA-C*03:02 | STFKCYGVS | FAFKCYGVS | 8370 | 424 |
| HLA-A*02:06 | VLYNSASFST | VLYNFAPFFA | 710 | 36 |
| HLA-B*15:03 | YSVLYNSASFS | YSVLYNFAPFF | 3170 | 164 |
| HLA-A*02:11 | VLYNSASFST | VLYNFAPFFA | 77 | 4 |
| HLA-A*02:01 | SVLYNSASFST | SVLYNFAPFFA | 5674 | 301 |
| HLA-A*11:01 | LYNSASFSTFK | LYNFAPFFAFK | 43 | 797 |
| HLA-A*03:02 | STFKCYGVSPTK | FAFKCYGVSPTK | 142 | 2541 |
| HLA-A*30:02 | SASFSTFKCY | FAPFFAFKCY | 71 | 1241 |
| HLA-A*02:11 | YSVLYNSASFST | YSVLYNFAPFFA | 7859 | 469 |
| HLA-C*14:02 | SFSTFKCY | PFFAFKCY | 386 | 6449 |
| HLA-A*30:01 | STFKCYGVS | FAFKCYGVS | 438 | 6895 |
| HLA-A*68:02 | FSTFKCYGV | FFAFKCYGV | 17 | 261 |
| HLA-A*32:01 | YNSASFSTF | YNFAPFFAF | 4333 | 294 |
| HLA-A*02:06 | FSTFKCYGV | FFAFKCYGV | 61 | 882 |
| HLA-A*33:03 | YNSASFSTFK | YNFAPFFAFK | 823 | 57 |
| HLA-A*11:01 | FSTFKCYGVSPTK | FFAFKCYGVSPTK | 426 | 5482 |
| HLA-A*02:06 | SVLYNSASFST | SVLYNFAPFFA | 4097 | 319 |
| HLA-B*18:01 | YNSASFSTF | YNFAPFFAF | 3495 | 284 |
| HLA-A*33:03 | NSASFSTFK | NFAPFFAFK | 181 | 16 |
| HLA-C*14:02 | FSTFKCYGV | FFAFKCYGV | 4043 | 371 |
| HLA-A*02:11 | VLYNSASFSTF | VLYNFAPFFAF | 2537 | 236 |
| HLA-A*11:01 | NSASFSTFK | NFAPFFAFK | 9 | 94 |
| HLA-A*29:02 | NSASFSTFKCY | NFAPFFAFKCY | 662 | 64 |
| HLA-A*29:02 | YNSASFSTFKCY | YNFAPFFAFKCY | 842 | 86 |
| HLA-A*02:01 | VLYNSASFSTF | VLYNFAPFFAF | 4199 | 455 |
| HLA-C*03:02 | YSVLYNSASFS | YSVLYNFAPFF | 2405 | 264 |
| HLA-A*29:02 | ASFSTFKCY | APFFAFKCY | 201 | 1830 |
| HLA-B*15:01 | VLYNSASFSTF | VLYNFAPFFAF | 50 | 420 |
| HLA-A*31:01 | YNSASFSTFK | YNFAPFFAFK | 607 | 73 |
| HLA-A*30:01 | LYNSASFST | LYNFAPFFA | 3467 | 433 |
| HLA-A*33:03 | SVLYNSASFSTFK | SVLYNFAPFFAFK | 2778 | 357 |
| HLA-B*35:01 | SASFSTFKCY | FAPFFAFKCY | 490 | 65 |
| HLA-A*30:01 | STFKCYGVSPTK | FAFKCYGVSPTK | 194 | 1428 |
| HLA-A*31:01 | NSASFSTFK | NFAPFFAFK | 293 | 47 |
| HLA-A*29:02 | LYNSASFSTF | LYNFAPFFAF | 387 | 63 |
| HLA-A*30:02 | VLYNSASFS | VLYNFAPFF | 2919 | 476 |
| HLA-A*31:01 | SVLYNSASFSTFK | SVLYNFAPFFAFK | 857 | 148 |
| HLA-A*23:01 | YNSASFSTF | YNFAPFFAF | 352 | 63 |
| HLA-A*03:01 | LYNSASFSTFK | LYNFAPFFAFK | 295 | 1614 |
| HLA-A*02:11 | SVLYNSASFS | SVLYNFAPFF | 2012 | 435 |
| HLA-A*03:01 | NSASFSTFK | NFAPFFAFK | 113 | 494 |
| HLA-B*35:01 | YNSASFSTF | YNFAPFFAF | 760 | 180 |
| HLA-A*03:02 | LYNSASFSTFK | LYNFAPFFAFK | 202 | 831 |
| HLA-A*02:01 | VLYNSASFS | VLYNFAPFF | 1548 | 377 |
| HLA-A*02:11 | VLYNSASFS | VLYNFAPFF | 258 | 63 |
| HLA-A*01:01 | SASFSTFKCY | FAPFFAFKCY | 461 | 1849 |
| HLA-A*11:01 | SASFSTFK | FAPFFAFK | 452 | 1795 |
| HLA-A*68:01 | SASFSTFK | FAPFFAFK | 1114 | 290 |
| HLA-B*15:01 | YSVLYNSASFSTF | YSVLYNFAPFFAF | 164 | 629 |
| HLA-A*03:02 | NSASFSTFK | NFAPFFAFK | 37 | 135 |
| HLA-A*30:02 | NSASFSTFKCY | NFAPFFAFKCY | 387 | 1395 |
| HLA-C*12:03 | SASFSTFKCY | FAPFFAFKCY | 498 | 139 |
| HLA-C*14:02 | LYNSASFSTF | LYNFAPFFAF | 20 | 69 |
| HLA-A*30:02 | VLYNSASFSTFKCY | VLYNFAPFFAFKCY | 272 | 923 |
| HLA-A*32:01 | SVLYNSASFSTF | SVLYNFAPFFAF | 1624 | 488 |
| HLA-A*31:01 | VLYNSASFSTFK | VLYNFAPFFAFK | 565 | 170 |
| HLA-A*68:01 | NSASFSTFK | NFAPFFAFK | 4 | 13 |
| HLA-A*31:01 | LYNSASFSTFK | LYNFAPFFAFK | 299 | 92 |
| HLA-A*11:01 | YNSASFSTFK | YNFAPFFAFK | 12 | 38 |
| HLA-A*24:02 | LYNSASFSTFK | LYNFAPFFAFK | 356 | 1101 |
| HLA-A*33:03 | LYNSASFSTFK | LYNFAPFFAFK | 1158 | 383 |
| HLA-A*24:02 | VLYNSASFSTF | VLYNFAPFFAF | 186 | 551 |
| HLA-A*03:01 | VLYNSASFSTFKC | VLYNFAPFFAFKC | 312 | 920 |
| HLA-A*02:06 | SASFSTFKCYGV | FAPFFAFKCYGV | 1416 | 481 |
| HLA-C*03:02 | YSVLYNSASFSTF | YSVLYNFAPFFAF | 113 | 329 |
| HLA-A*68:01 | FSTFKCYGVSPTK | FFAFKCYGVSPTK | 288 | 832 |
| HLA-B*58:01 | YSVLYNSASFSTF | YSVLYNFAPFFAF | 429 | 1160 |
| HLA-A*29:02 | LYNSASFSTFKCY | LYNFAPFFAFKCY | 297 | 115 |
| HLA-C*12:02 | SASFSTFKCY | FAPFFAFKCY | 710 | 275 |
| HLA-B*15:03 | VLYNSASFSTF | VLYNFAPFFAF | 38 | 96 |
| HLA-A*68:01 | NSASFSTFKC | NFAPFFAFKC | 281 | 702 |
| HLA-A*03:01 | VLYNSASFSTFK | VLYNFAPFFAFK | 7 | 17 |
| HLA-A*03:01 | YNSASFSTFK | YNFAPFFAFK | 80 | 194 |
| HLA-A*11:01 | VLYNSASFSTFK | VLYNFAPFFAFK | 22 | 51 |
| HLA-B*15:01 | SVLYNSASFSTF | SVLYNFAPFFAF | 221 | 509 |
| HLA-B*15:01 | LYNSASFSTF | LYNFAPFFAF | 468 | 1064 |
| HLA-B*15:01 | SASFSTFKCY | FAPFFAFKCY | 256 | 561 |
| HLA-C*03:02 | SASFSTFKCY | FAPFFAFKCY | 212 | 97 |
| HLA-A*68:01 | YSVLYNSASFSTFK | YSVLYNFAPFFAFK | 144 | 67 |
| HLA-A*30:01 | TFKCYGVSPTK | AFKCYGVSPTK | 319 | 155 |
| HLA-A*02:01 | FSTFKCYGV | FFAFKCYGV | 457 | 926 |
D405N
Reference YGVSPTKLNDLCFTNVYADSFVIRGDEVRQIAPGQTGKIADYNYKLPDDFT
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated YGVSPTKLNDLCFTNVYADSFVIRGNEVSQIAPGQTGNIADYNYKLPDDFT
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-A*68:01 | FVIRGDEVR | FVIRGNEVS | 23 | 15265 |
| HLA-A*68:01 | SFVIRGDEVR | SFVIRGNEVS | 186 | 29639 |
| HLA-A*68:01 | DSFVIRGDEVR | DSFVIRGNEVS | 220 | 29286 |
| HLA-A*33:03 | FVIRGDEVR | FVIRGNEVS | 320 | 29448 |
| HLA-A*33:03 | SFVIRGDEVR | SFVIRGNEVS | 459 | 31986 |
| HLA-C*14:02 | SFVIRGDEV | SFVIRGNEV | 216 | 46 |
| HLA-C*06:02 | IRGDEVRQI | IRGNEVSQI | 147 | 642 |
R408S
Reference SPTKLNDLCFTNVYADSFVIRGDEVRQIAPGQTGKIADYNYKLPDDFTGCV
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated SPTKLNDLCFTNVYADSFVIRGNEVSQIAPGQTGNIADYNYKLPDDFTGCV
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-A*68:01 | FVIRGDEVR | FVIRGNEVS | 23 | 15265 |
| HLA-A*03:01 | RQIAPGQTGK | SQIAPGQTGN | 49 | 32429 |
| HLA-A*11:01 | RQIAPGQTGK | SQIAPGQTGN | 103 | 32387 |
| HLA-A*03:02 | RQIAPGQTGK | SQIAPGQTGN | 123 | 32767 |
| HLA-A*68:01 | EVRQIAPGQTGK | EVSQIAPGQTGN | 144 | 32361 |
| HLA-A*68:01 | SFVIRGDEVR | SFVIRGNEVS | 186 | 29639 |
| HLA-A*68:01 | DSFVIRGDEVR | DSFVIRGNEVS | 220 | 29286 |
| HLA-A*33:03 | FVIRGDEVR | FVIRGNEVS | 320 | 29448 |
| HLA-A*33:03 | SFVIRGDEVR | SFVIRGNEVS | 459 | 31986 |
| HLA-A*30:01 | RQIAPGQTGK | SQIAPGQTGN | 275 | 18394 |
| HLA-C*06:02 | IRGDEVRQI | IRGNEVSQI | 147 | 642 |
| HLA-B*15:03 | RQIAPGQT | SQIAPGQT | 478 | 1764 |
| HLA-B*15:03 | RQIAPGQTGKIADY | SQIAPGQTGNIADY | 17 | 55 |
| HLA-A*30:02 | RQIAPGQTGKIADY | SQIAPGQTGNIADY | 324 | 951 |
| HLA-B*15:03 | RQIAPGQTGKI | SQIAPGQTGNI | 236 | 691 |
| HLA-B*15:03 | RQIAPGQTG | SQIAPGQTG | 39 | 107 |
K417N
Reference FTNVYADSFVIRGDEVRQIAPGQTGKIADYNYKLPDDFTGCVIAWNSNNLD
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated FTNVYADSFVIRGNEVSQIAPGQTGNIADYNYKLPDDFTGCVIAWNSNKLD
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-A*03:01 | RQIAPGQTGK | SQIAPGQTGN | 49 | 32429 |
| HLA-A*11:01 | RQIAPGQTGK | SQIAPGQTGN | 103 | 32387 |
| HLA-A*03:02 | RQIAPGQTGK | SQIAPGQTGN | 123 | 32767 |
| HLA-A*68:01 | EVRQIAPGQTGK | EVSQIAPGQTGN | 144 | 32361 |
| HLA-A*68:01 | QIAPGQTGK | QIAPGQTGN | 136 | 27997 |
| HLA-A*11:01 | QIAPGQTGK | QIAPGQTGN | 165 | 32767 |
| HLA-A*03:02 | QIAPGQTGK | QIAPGQTGN | 278 | 32767 |
| HLA-A*03:01 | QIAPGQTGK | QIAPGQTGN | 296 | 32767 |
| HLA-A*32:01 | KIADYNYKL | NIADYNYKL | 58 | 4480 |
| HLA-A*30:01 | RQIAPGQTGK | SQIAPGQTGN | 275 | 18394 |
| HLA-A*68:02 | KIADYNYKL | NIADYNYKL | 1274 | 39 |
| HLA-A*03:02 | KIADYNYK | NIADYNYK | 476 | 6850 |
| HLA-A*02:06 | KIADYNYKL | NIADYNYKL | 20 | 101 |
| HLA-A*02:01 | KIADYNYKL | NIADYNYKL | 23 | 110 |
| HLA-C*17:01 | KIADYNYKL | NIADYNYKL | 155 | 669 |
| HLA-A*30:01 | TGKIADYNYK | TGNIADYNYK | 292 | 1164 |
| HLA-A*02:11 | KIADYNYKL | NIADYNYKL | 5 | 19 |
| HLA-B*15:03 | RQIAPGQTGKIADY | SQIAPGQTGNIADY | 17 | 55 |
| HLA-A*30:02 | RQIAPGQTGKIADY | SQIAPGQTGNIADY | 324 | 951 |
| HLA-B*15:03 | RQIAPGQTGKI | SQIAPGQTGNI | 236 | 691 |
| HLA-A*68:01 | TGKIADYNYK | TGNIADYNYK | 957 | 343 |
| HLA-A*01:01 | QTGKIADYNY | QTGNIADYNY | 580 | 270 |
N440K
Reference TGKIADYNYKLPDDFTGCVIAWNSNNLDSKVGGNYNYLYRLFRKSNLKPFE
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated TGNIADYNYKLPDDFTGCVIAWNSNKLDSKVGGNYNYRYRLFRKSNLKPFE
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-A*11:01 | CVIAWNSNN | CVIAWNSNK | 24486 | 26 |
| HLA-A*68:01 | CVIAWNSNN | CVIAWNSNK | 11802 | 16 |
| HLA-A*68:01 | FTGCVIAWNSNN | FTGCVIAWNSNK | 25655 | 59 |
| HLA-A*03:01 | CVIAWNSNN | CVIAWNSNK | 24643 | 61 |
| HLA-A*03:02 | CVIAWNSNN | CVIAWNSNK | 22621 | 61 |
| HLA-A*11:01 | GCVIAWNSNN | GCVIAWNSNK | 32767 | 94 |
| HLA-A*03:01 | GCVIAWNSNN | GCVIAWNSNK | 32767 | 128 |
| HLA-A*11:01 | TGCVIAWNSNN | TGCVIAWNSNK | 32767 | 153 |
| HLA-A*68:01 | TGCVIAWNSNN | TGCVIAWNSNK | 32767 | 180 |
| HLA-A*68:01 | DFTGCVIAWNSNN | DFTGCVIAWNSNK | 32767 | 255 |
| HLA-A*03:02 | GCVIAWNSNN | GCVIAWNSNK | 32767 | 277 |
| HLA-A*31:01 | NLDSKVGGNYNYL | KLDSKVGGNYNYR | 31768 | 301 |
| HLA-A*11:01 | FTGCVIAWNSNN | FTGCVIAWNSNK | 32767 | 408 |
| HLA-A*31:01 | CVIAWNSNN | CVIAWNSNK | 18631 | 279 |
| HLA-A*33:03 | CVIAWNSNN | CVIAWNSNK | 20952 | 493 |
| HLA-A*30:01 | CVIAWNSNN | CVIAWNSNK | 11396 | 467 |
| HLA-A*01:01 | NLDSKVGGNYNYLY | KLDSKVGGNYNYRY | 193 | 2088 |
| HLA-A*30:02 | NLDSKVGGNYNY | KLDSKVGGNYNY | 3839 | 409 |
| HLA-A*30:02 | NLDSKVGGNY | KLDSKVGGNY | 422 | 52 |
| HLA-B*15:01 | NLDSKVGGNY | KLDSKVGGNY | 1404 | 179 |
| HLA-A*01:01 | NLDSKVGGNY | KLDSKVGGNY | 371 | 819 |
| HLA-A*02:11 | VIAWNSNNL | VIAWNSNKL | 269 | 586 |
| HLA-A*68:02 | FTGCVIAWNSNNL | FTGCVIAWNSNKL | 461 | 960 |
L452R
Reference DDFTGCVIAWNSNNLDSKVGGNYNYLYRLFRKSNLKPFERDISTEIYQAGS
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated DDFTGCVIAWNSNKLDSKVGGNYNYRYRLFRKSNLKPFERDISTEIYQAGN
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-A*31:01 | KVGGNYNYL | KVGGNYNYR | 5813 | 13 |
| HLA-A*02:11 | YLYRLFRKSNL | YRYRLFRKSNL | 82 | 22451 |
| HLA-A*68:01 | SKVGGNYNYL | SKVGGNYNYR | 24692 | 119 |
| HLA-B*27:05 | YLYRLFRK | YRYRLFRK | 9806 | 50 |
| HLA-A*02:11 | YLYRLFRKS | YRYRLFRKS | 179 | 28031 |
| HLA-A*02:02 | YLYRLFRKSNL | YRYRLFRKSNL | 188 | 23354 |
| HLA-A*02:11 | KVGGNYNYL | KVGGNYNYR | 111 | 13729 |
| HLA-B*27:05 | YLYRLFRKS | YRYRLFRKS | 13862 | 117 |
| HLA-A*31:01 | NLDSKVGGNYNYL | KLDSKVGGNYNYR | 31768 | 301 |
| HLA-A*33:03 | DSKVGGNYNYL | DSKVGGNYNYR | 30679 | 292 |
| HLA-B*27:05 | YLYRLFRKSNLK | YRYRLFRKSNLK | 2218 | 22 |
| HLA-A*31:01 | SKVGGNYNYL | SKVGGNYNYR | 21677 | 233 |
| HLA-A*03:01 | YLYRLFRK | YRYRLFRK | 204 | 18425 |
| HLA-A*68:01 | KVGGNYNYL | KVGGNYNYR | 25432 | 286 |
| HLA-A*33:03 | KVGGNYNYL | KVGGNYNYR | 22954 | 269 |
| HLA-A*11:01 | KVGGNYNYL | KVGGNYNYR | 16150 | 194 |
| HLA-A*02:02 | KVGGNYNYL | KVGGNYNYR | 279 | 23086 |
| HLA-A*68:01 | DSKVGGNYNYL | DSKVGGNYNYR | 28915 | 378 |
| HLA-A*03:02 | YLYRLFRK | YRYRLFRK | 327 | 22983 |
| HLA-A*02:01 | YLYRLFRKSNL | YRYRLFRKSNL | 471 | 28211 |
| HLA-A*33:03 | SKVGGNYNYL | SKVGGNYNYR | 27973 | 469 |
| HLA-A*03:02 | KVGGNYNYL | KVGGNYNYR | 12848 | 234 |
| HLA-A*02:06 | KVGGNYNYL | KVGGNYNYR | 452 | 18600 |
| HLA-B*27:05 | YLYRLFRKSNLKP | YRYRLFRKSNLKP | 13004 | 347 |
| HLA-B*27:05 | YLYRLFRKSNLKPF | YRYRLFRKSNLKPF | 7101 | 214 |
| HLA-B*27:05 | YLYRLFRKSNL | YRYRLFRKSNL | 1360 | 48 |
| HLA-B*15:01 | YLYRLFRKSNLKPF | YRYRLFRKSNLKPF | 476 | 9205 |
| HLA-B*08:01 | YLYRLFRKSNL | YRYRLFRKSNL | 160 | 1946 |
| HLA-A*30:01 | LYRLFRKSN | RYRLFRKSN | 1444 | 120 |
| HLA-A*30:01 | LYRLFRKSNL | RYRLFRKSNL | 1104 | 93 |
| HLA-A*02:11 | KVGGNYNYLYRL | KVGGNYNYRYRL | 274 | 3087 |
| HLA-A*30:01 | LYRLFRKSNLKP | RYRLFRKSNLKP | 3059 | 277 |
| HLA-A*01:01 | NLDSKVGGNYNYLY | KLDSKVGGNYNYRY | 193 | 2088 |
| HLA-A*30:01 | LYRLFRKSNLK | RYRLFRKSNLK | 192 | 21 |
| HLA-A*03:01 | LYRLFRKSNLKP | RYRLFRKSNLKP | 3564 | 430 |
| HLA-A*30:01 | NYLYRLFRK | NYRYRLFRK | 512 | 64 |
| HLA-B*27:05 | LYRLFRKSNL | RYRLFRKSNL | 932 | 123 |
| HLA-A*23:01 | GNYNYLYRLF | GNYNYRYRLF | 181 | 1119 |
| HLA-A*23:01 | NYNYLYRLF | NYNYRYRLF | 20 | 119 |
| HLA-A*23:01 | KVGGNYNYLYRLF | KVGGNYNYRYRLF | 410 | 2404 |
| HLA-A*23:01 | VGGNYNYLYRLF | VGGNYNYRYRLF | 222 | 1236 |
| HLA-A*29:02 | VGGNYNYLY | VGGNYNYRY | 94 | 518 |
| HLA-A*31:01 | LYRLFRKSNLK | RYRLFRKSNLK | 356 | 67 |
| HLA-A*30:01 | YLYRLFRKSNLK | YRYRLFRKSNLK | 1354 | 258 |
| HLA-A*24:02 | NYNYLYRLF | NYNYRYRLF | 21 | 108 |
| HLA-A*29:02 | KVGGNYNYLY | KVGGNYNYRY | 100 | 505 |
| HLA-A*24:02 | GNYNYLYRLF | GNYNYRYRLF | 325 | 1597 |
| HLA-A*24:02 | VGGNYNYLYRLF | VGGNYNYRYRLF | 391 | 1596 |
| HLA-A*03:01 | LYRLFRKSNLK | RYRLFRKSNLK | 65 | 18 |
| HLA-A*03:02 | LYRLFRKSNLK | RYRLFRKSNLK | 242 | 72 |
| HLA-A*30:01 | YNYLYRLFRK | YNYRYRLFRK | 379 | 125 |
| HLA-A*11:01 | LYRLFRKSNLK | RYRLFRKSNLK | 952 | 401 |
| HLA-B*15:03 | SKVGGNYNYLY | SKVGGNYNYRY | 313 | 720 |
| HLA-A*30:02 | VGGNYNYLY | VGGNYNYRY | 45 | 93 |
477-478 ST->NK
Reference LYRLFRKSNLKPFERDISTEIYQAGSTPCNGVEGFNCYFPLQSYGFQPTNGV
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated RYRLFRKSNLKPFERDISTEIYQAGNKPCNGVAGVNCYFPLQSYGFRPTYGV
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-A*11:01 | STEIYQAGST | STEIYQAGNK | 30358 | 67 |
| HLA-A*68:01 | STEIYQAGST | STEIYQAGNK | 22142 | 157 |
| HLA-A*02:02 | YQAGSTPCNGVEGF | YQAGNKPCNGVAGV | 9447 | 68 |
| HLA-A*03:01 | STEIYQAGST | STEIYQAGNK | 32241 | 308 |
| HLA-A*02:11 | YQAGSTPCNGVEGF | YQAGNKPCNGVAGV | 17568 | 184 |
| HLA-B*35:01 | TPCNGVEGF | KPCNGVAGV | 319 | 21824 |
| HLA-A*02:06 | YQAGSTPCNGVEGF | YQAGNKPCNGVAGV | 5118 | 87 |
| HLA-B*15:03 | YQAGSTPCNGVEGF | YQAGNKPCNGVAGV | 217 | 12581 |
| HLA-B*15:01 | YQAGSTPCNGVEGF | YQAGNKPCNGVAGV | 408 | 12442 |
| HLA-A*02:11 | YQAGSTPCNGV | YQAGNKPCNGV | 173 | 678 |
| HLA-B*50:01 | TEIYQAGSTP | TEIYQAGNKP | 464 | 1807 |
| HLA-A*02:06 | YQAGSTPCNGV | YQAGNKPCNGV | 39 | 146 |
| HLA-A*02:01 | YQAGSTPCNGV | YQAGNKPCNGV | 420 | 1490 |
| HLA-C*14:02 | IYQAGSTPC | IYQAGNKPC | 134 | 459 |
| HLA-A*02:02 | YQAGSTPCNGV | YQAGNKPCNGV | 107 | 233 |
E484A
Reference SNLKPFERDISTEIYQAGSTPCNGVEGFNCYFPLQSYGFQPTNGVGYQPYR
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated SNLKPFERDISTEIYQAGNKPCNGVAGVNCYFPLQSYGFRPTYGVGHQPYR
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-B*40:02 | VEGFNCYFPL | VAGVNCYFPL | 31 | 19248 |
| HLA-B*40:01 | VEGFNCYFPL | VAGVNCYFPL | 76 | 26041 |
| HLA-A*02:02 | YQAGSTPCNGVEGF | YQAGNKPCNGVAGV | 9447 | 68 |
| HLA-A*02:11 | YQAGSTPCNGVEGF | YQAGNKPCNGVAGV | 17568 | 184 |
| HLA-B*35:01 | TPCNGVEGF | KPCNGVAGV | 319 | 21824 |
| HLA-A*02:06 | YQAGSTPCNGVEGF | YQAGNKPCNGVAGV | 5118 | 87 |
| HLA-B*15:03 | YQAGSTPCNGVEGF | YQAGNKPCNGVAGV | 217 | 12581 |
| HLA-A*68:02 | EGFNCYFPL | AGVNCYFPL | 117 | 4903 |
| HLA-B*15:01 | GVEGFNCYF | GVAGVNCYF | 3125 | 98 |
| HLA-B*15:01 | YQAGSTPCNGVEGF | YQAGNKPCNGVAGV | 408 | 12442 |
| HLA-B*15:03 | GVEGFNCYF | GVAGVNCYF | 1657 | 235 |
| HLA-A*30:02 | EGFNCYFPLQSY | AGVNCYFPLQSY | 1806 | 447 |
| HLA-A*02:02 | GVEGFNCYFPL | GVAGVNCYFPL | 666 | 171 |
| HLA-B*35:01 | NGVEGFNCY | NGVAGVNCY | 346 | 866 |
F486V
Reference LKPFERDISTEIYQAGSTPCNGVEGFNCYFPLQSYGFQPTNGVGYQPYRVV
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated LKPFERDISTEIYQAGNKPCNGVAGVNCYFPLQSYGFRPTYGVGHQPYRVV
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-B*40:02 | VEGFNCYFPL | VAGVNCYFPL | 31 | 19248 |
| HLA-B*40:01 | VEGFNCYFPL | VAGVNCYFPL | 76 | 26041 |
| HLA-A*02:02 | YQAGSTPCNGVEGF | YQAGNKPCNGVAGV | 9447 | 68 |
| HLA-A*02:11 | YQAGSTPCNGVEGF | YQAGNKPCNGVAGV | 17568 | 184 |
| HLA-B*35:01 | TPCNGVEGF | KPCNGVAGV | 319 | 21824 |
| HLA-A*02:06 | YQAGSTPCNGVEGF | YQAGNKPCNGVAGV | 5118 | 87 |
| HLA-B*15:03 | YQAGSTPCNGVEGF | YQAGNKPCNGVAGV | 217 | 12581 |
| HLA-A*68:02 | EGFNCYFPL | AGVNCYFPL | 117 | 4903 |
| HLA-B*15:01 | GVEGFNCYF | GVAGVNCYF | 3125 | 98 |
| HLA-B*15:01 | YQAGSTPCNGVEGF | YQAGNKPCNGVAGV | 408 | 12442 |
| HLA-C*03:02 | FNCYFPLQSY | VNCYFPLQSY | 225 | 1964 |
| HLA-B*15:03 | GVEGFNCYF | GVAGVNCYF | 1657 | 235 |
| HLA-A*30:02 | EGFNCYFPLQSY | AGVNCYFPLQSY | 1806 | 447 |
| HLA-A*02:02 | GVEGFNCYFPL | GVAGVNCYFPL | 666 | 171 |
| HLA-A*29:02 | GFNCYFPLQSY | GVNCYFPLQSY | 191 | 742 |
| HLA-A*29:02 | FNCYFPLQSY | VNCYFPLQSY | 73 | 227 |
| HLA-B*35:01 | NGVEGFNCY | NGVAGVNCY | 346 | 866 |
| HLA-B*15:03 | FNCYFPLQSY | VNCYFPLQSY | 101 | 240 |
Q498R
Reference YQAGSTPCNGVEGFNCYFPLQSYGFQPTNGVGYQPYRVVVLSFELLHAPAT
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated YQAGNKPCNGVAGVNCYFPLQSYGFRPTYGVGHQPYRVVVLSFELLHAPAT
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-C*03:02 | QSYGFQPTN | QSYGFRPTY | 12903 | 18 |
| HLA-B*15:01 | LQSYGFQPTN | LQSYGFRPTY | 6135 | 9 |
| HLA-C*16:01 | QSYGFQPTN | QSYGFRPTY | 10868 | 16 |
| HLA-A*29:02 | QSYGFQPTN | QSYGFRPTY | 26152 | 43 |
| HLA-B*15:03 | QSYGFQPTN | QSYGFRPTY | 6543 | 11 |
| HLA-B*15:03 | LQSYGFQPTN | LQSYGFRPTY | 2045 | 4 |
| HLA-C*12:03 | QSYGFQPTN | QSYGFRPTY | 12696 | 25 |
| HLA-A*29:02 | YFPLQSYGFQPTN | YFPLQSYGFRPTY | 19514 | 47 |
| HLA-B*15:03 | PLQSYGFQPTN | PLQSYGFRPTY | 22802 | 60 |
| HLA-B*35:01 | FPLQSYGFQPTN | FPLQSYGFRPTY | 8715 | 27 |
| HLA-A*33:03 | YFPLQSYGFQ | YFPLQSYGFR | 11148 | 35 |
| HLA-C*14:02 | SYGFQPTN | SYGFRPTY | 20230 | 66 |
| HLA-A*31:01 | CYFPLQSYGFQ | CYFPLQSYGFR | 8535 | 34 |
| HLA-C*03:02 | FPLQSYGFQPTN | FPLQSYGFRPTY | 23840 | 111 |
| HLA-C*14:02 | YFPLQSYGFQPTN | YFPLQSYGFRPTY | 18230 | 85 |
| HLA-A*33:03 | CYFPLQSYGFQ | CYFPLQSYGFR | 11311 | 54 |
| HLA-C*03:02 | LQSYGFQPTN | LQSYGFRPTY | 23421 | 125 |
| HLA-B*15:01 | FQPTNGVGY | FRPTYGVGH | 166 | 29043 |
| HLA-A*30:02 | QSYGFQPTN | QSYGFRPTY | 5297 | 32 |
| HLA-A*33:03 | FPLQSYGFQ | FPLQSYGFR | 23527 | 146 |
| HLA-B*15:03 | FPLQSYGFQPTN | FPLQSYGFRPTY | 20159 | 126 |
| HLA-A*31:01 | YFPLQSYGFQ | YFPLQSYGFR | 16290 | 103 |
| HLA-B*35:01 | QSYGFQPTN | QSYGFRPTY | 24615 | 156 |
| HLA-C*03:02 | YFPLQSYGFQPTN | YFPLQSYGFRPTY | 27639 | 176 |
| HLA-C*16:01 | LQSYGFQPTN | LQSYGFRPTY | 27149 | 187 |
| HLA-A*30:02 | LQSYGFQPTN | LQSYGFRPTY | 7508 | 54 |
| HLA-B*15:01 | QSYGFQPTN | QSYGFRPTY | 21529 | 157 |
| HLA-C*12:02 | QSYGFQPTN | QSYGFRPTY | 21029 | 169 |
| HLA-B*15:03 | YFPLQSYGFQPTN | YFPLQSYGFRPTY | 22168 | 189 |
| HLA-C*16:01 | FPLQSYGFQPTN | FPLQSYGFRPTY | 27733 | 237 |
| HLA-A*29:02 | CYFPLQSYGFQPTN | CYFPLQSYGFRPTY | 27182 | 240 |
| HLA-A*29:02 | LQSYGFQPTN | LQSYGFRPTY | 28458 | 265 |
| HLA-B*15:03 | FQPTNGVGY | FRPTYGVGH | 63 | 6390 |
| HLA-A*29:02 | GFQPTNGVGY | GFRPTYGVGH | 188 | 18753 |
| HLA-C*16:01 | YFPLQSYGFQPTN | YFPLQSYGFRPTY | 29358 | 306 |
| HLA-C*12:03 | FPLQSYGFQPTN | FPLQSYGFRPTY | 27893 | 295 |
| HLA-C*14:02 | QSYGFQPTN | QSYGFRPTY | 25941 | 277 |
| HLA-A*31:01 | NCYFPLQSYGFQ | NCYFPLQSYGFR | 21321 | 230 |
| HLA-A*33:03 | NCYFPLQSYGFQ | NCYFPLQSYGFR | 22134 | 257 |
| HLA-C*12:03 | LQSYGFQPTN | LQSYGFRPTY | 26641 | 325 |
| HLA-B*15:03 | LQSYGFQPTNG | LQSYGFRPTYG | 6387 | 82 |
| HLA-A*32:01 | QSYGFQPTN | QSYGFRPTY | 16619 | 226 |
| HLA-A*11:01 | QSYGFQPTN | QSYGFRPTY | 22068 | 301 |
| HLA-A*68:01 | FPLQSYGFQ | FPLQSYGFR | 17691 | 247 |
| HLA-C*02:02 | QSYGFQPTN | QSYGFRPTY | 26598 | 412 |
| HLA-C*02:10 | QSYGFQPTN | QSYGFRPTY | 26598 | 412 |
| HLA-A*68:01 | YFPLQSYGFQ | YFPLQSYGFR | 14480 | 278 |
| HLA-C*14:02 | CYFPLQSYGFQPTN | CYFPLQSYGFRPTY | 24158 | 500 |
| HLA-A*30:02 | YFPLQSYGFQPTN | YFPLQSYGFRPTY | 18743 | 390 |
| HLA-B*15:01 | LQSYGFQPTNGVGY | LQSYGFRPTYGVGH | 180 | 8335 |
| HLA-B*15:01 | FQPTNGVGYQPY | FRPTYGVGHQPY | 290 | 11634 |
| HLA-B*15:03 | YGFQPTNGVGY | YGFRPTYGVGH | 368 | 10641 |
| HLA-A*30:02 | GFQPTNGVGY | GFRPTYGVGH | 169 | 4730 |
| HLA-A*30:02 | QSYGFQPTNGVGY | QSYGFRPTYGVGH | 258 | 7114 |
| HLA-B*15:03 | LQSYGFQPTNGVGY | LQSYGFRPTYGVGH | 113 | 2917 |
| HLA-B*53:01 | FPLQSYGFQPTN | FPLQSYGFRPTY | 8451 | 351 |
| HLA-C*07:02 | FQPTNGVGYQPY | FRPTYGVGHQPY | 6169 | 356 |
| HLA-B*57:01 | QSYGFQPTN | QSYGFRPTY | 4070 | 293 |
| HLA-A*30:01 | QSYGFQPTN | QSYGFRPTY | 3720 | 308 |
| HLA-B*57:01 | LQSYGFQPTN | LQSYGFRPTY | 3839 | 375 |
| HLA-C*14:02 | FQPTNGVGYQPY | FRPTYGVGHQPY | 3107 | 366 |
| HLA-B*15:03 | FQPTNGVGYQPY | FRPTYGVGHQPY | 183 | 1236 |
| HLA-A*31:01 | QPTNGVGYQPYR | RPTYGVGHQPYR | 3100 | 478 |
| HLA-B*58:01 | QSYGFQPTN | QSYGFRPTY | 1532 | 249 |
| HLA-A*23:01 | SYGFQPTNGV | SYGFRPTYGV | 2697 | 483 |
| HLA-A*02:11 | YGFQPTNGV | YGFRPTYGV | 307 | 59 |
| HLA-B*15:03 | LQSYGFQPTNGV | LQSYGFRPTYGV | 1971 | 442 |
| HLA-A*02:02 | YGFQPTNGV | YGFRPTYGV | 868 | 281 |
| HLA-A*31:01 | GFQPTNGVGYQPYR | GFRPTYGVGHQPYR | 616 | 201 |
| HLA-A*02:06 | YGFQPTNGV | YGFRPTYGV | 174 | 61 |
| HLA-A*68:02 | YGFQPTNGV | YGFRPTYGV | 73 | 26 |
| HLA-C*12:03 | YGFQPTNGV | YGFRPTYGV | 127 | 355 |
| HLA-A*02:06 | QSYGFQPTNGV | QSYGFRPTYGV | 1121 | 425 |
| HLA-A*68:02 | QSYGFQPTNGV | QSYGFRPTYGV | 122 | 47 |
| HLA-A*02:06 | LQSYGFQPT | LQSYGFRPT | 339 | 871 |
N501Y
Reference GSTPCNGVEGFNCYFPLQSYGFQPTNGVGYQPYRVVVLSFELLHAPATVCG
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated GNKPCNGVAGVNCYFPLQSYGFRPTYGVGHQPYRVVVLSFELLHAPATVCG
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-C*03:02 | QSYGFQPTN | QSYGFRPTY | 12903 | 18 |
| HLA-B*15:01 | LQSYGFQPTN | LQSYGFRPTY | 6135 | 9 |
| HLA-C*16:01 | QSYGFQPTN | QSYGFRPTY | 10868 | 16 |
| HLA-A*29:02 | QSYGFQPTN | QSYGFRPTY | 26152 | 43 |
| HLA-B*15:03 | QSYGFQPTN | QSYGFRPTY | 6543 | 11 |
| HLA-B*15:03 | LQSYGFQPTN | LQSYGFRPTY | 2045 | 4 |
| HLA-C*12:03 | QSYGFQPTN | QSYGFRPTY | 12696 | 25 |
| HLA-A*29:02 | YFPLQSYGFQPTN | YFPLQSYGFRPTY | 19514 | 47 |
| HLA-B*15:03 | PLQSYGFQPTN | PLQSYGFRPTY | 22802 | 60 |
| HLA-C*14:02 | TNGVGYQPY | TYGVGHQPY | 5468 | 16 |
| HLA-B*35:01 | FPLQSYGFQPTN | FPLQSYGFRPTY | 8715 | 27 |
| HLA-C*14:02 | SYGFQPTN | SYGFRPTY | 20230 | 66 |
| HLA-C*03:02 | FPLQSYGFQPTN | FPLQSYGFRPTY | 23840 | 111 |
| HLA-C*14:02 | YFPLQSYGFQPTN | YFPLQSYGFRPTY | 18230 | 85 |
| HLA-C*03:02 | LQSYGFQPTN | LQSYGFRPTY | 23421 | 125 |
| HLA-B*15:01 | FQPTNGVGY | FRPTYGVGH | 166 | 29043 |
| HLA-A*30:02 | QSYGFQPTN | QSYGFRPTY | 5297 | 32 |
| HLA-B*15:03 | FPLQSYGFQPTN | FPLQSYGFRPTY | 20159 | 126 |
| HLA-B*35:01 | QSYGFQPTN | QSYGFRPTY | 24615 | 156 |
| HLA-C*03:02 | YFPLQSYGFQPTN | YFPLQSYGFRPTY | 27639 | 176 |
| HLA-C*16:01 | LQSYGFQPTN | LQSYGFRPTY | 27149 | 187 |
| HLA-A*30:02 | LQSYGFQPTN | LQSYGFRPTY | 7508 | 54 |
| HLA-B*15:01 | QSYGFQPTN | QSYGFRPTY | 21529 | 157 |
| HLA-C*12:02 | QSYGFQPTN | QSYGFRPTY | 21029 | 169 |
| HLA-B*15:03 | YFPLQSYGFQPTN | YFPLQSYGFRPTY | 22168 | 189 |
| HLA-C*16:01 | FPLQSYGFQPTN | FPLQSYGFRPTY | 27733 | 237 |
| HLA-A*29:02 | CYFPLQSYGFQPTN | CYFPLQSYGFRPTY | 27182 | 240 |
| HLA-A*29:02 | LQSYGFQPTN | LQSYGFRPTY | 28458 | 265 |
| HLA-B*15:03 | FQPTNGVGY | FRPTYGVGH | 63 | 6390 |
| HLA-A*29:02 | GFQPTNGVGY | GFRPTYGVGH | 188 | 18753 |
| HLA-C*16:01 | YFPLQSYGFQPTN | YFPLQSYGFRPTY | 29358 | 306 |
| HLA-C*12:03 | FPLQSYGFQPTN | FPLQSYGFRPTY | 27893 | 295 |
| HLA-C*14:02 | QSYGFQPTN | QSYGFRPTY | 25941 | 277 |
| HLA-C*12:03 | LQSYGFQPTN | LQSYGFRPTY | 26641 | 325 |
| HLA-B*15:03 | LQSYGFQPTNG | LQSYGFRPTYG | 6387 | 82 |
| HLA-A*32:01 | QSYGFQPTN | QSYGFRPTY | 16619 | 226 |
| HLA-A*11:01 | QSYGFQPTN | QSYGFRPTY | 22068 | 301 |
| HLA-C*02:02 | QSYGFQPTN | QSYGFRPTY | 26598 | 412 |
| HLA-C*02:10 | QSYGFQPTN | QSYGFRPTY | 26598 | 412 |
| HLA-C*14:02 | CYFPLQSYGFQPTN | CYFPLQSYGFRPTY | 24158 | 500 |
| HLA-A*30:02 | YFPLQSYGFQPTN | YFPLQSYGFRPTY | 18743 | 390 |
| HLA-B*15:01 | LQSYGFQPTNGVGY | LQSYGFRPTYGVGH | 180 | 8335 |
| HLA-B*15:01 | FQPTNGVGYQPY | FRPTYGVGHQPY | 290 | 11634 |
| HLA-C*03:02 | NGVGYQPY | YGVGHQPY | 10463 | 322 |
| HLA-B*15:03 | YGFQPTNGVGY | YGFRPTYGVGH | 368 | 10641 |
| HLA-A*30:02 | GFQPTNGVGY | GFRPTYGVGH | 169 | 4730 |
| HLA-A*30:02 | QSYGFQPTNGVGY | QSYGFRPTYGVGH | 258 | 7114 |
| HLA-B*15:03 | LQSYGFQPTNGVGY | LQSYGFRPTYGVGH | 113 | 2917 |
| HLA-B*53:01 | FPLQSYGFQPTN | FPLQSYGFRPTY | 8451 | 351 |
| HLA-C*16:01 | NGVGYQPY | YGVGHQPY | 8669 | 488 |
| HLA-C*07:02 | FQPTNGVGYQPY | FRPTYGVGHQPY | 6169 | 356 |
| HLA-B*57:01 | QSYGFQPTN | QSYGFRPTY | 4070 | 293 |
| HLA-A*30:01 | QSYGFQPTN | QSYGFRPTY | 3720 | 308 |
| HLA-A*02:11 | NGVGYQPYRV | YGVGHQPYRV | 5989 | 500 |
| HLA-B*15:03 | NGVGYQPY | YGVGHQPY | 4677 | 418 |
| HLA-B*57:01 | LQSYGFQPTN | LQSYGFRPTY | 3839 | 375 |
| HLA-C*14:02 | FQPTNGVGYQPY | FRPTYGVGHQPY | 3107 | 366 |
| HLA-A*29:02 | TNGVGYQPY | TYGVGHQPY | 2031 | 243 |
| HLA-A*31:01 | TNGVGYQPYR | TYGVGHQPYR | 811 | 103 |
| HLA-A*33:03 | TNGVGYQPYR | TYGVGHQPYR | 573 | 79 |
| HLA-B*15:03 | FQPTNGVGYQPY | FRPTYGVGHQPY | 183 | 1236 |
| HLA-A*31:01 | QPTNGVGYQPYR | RPTYGVGHQPYR | 3100 | 478 |
| HLA-B*58:01 | QSYGFQPTN | QSYGFRPTY | 1532 | 249 |
| HLA-A*23:01 | SYGFQPTNGV | SYGFRPTYGV | 2697 | 483 |
| HLA-A*02:11 | YGFQPTNGV | YGFRPTYGV | 307 | 59 |
| HLA-B*15:03 | LQSYGFQPTNGV | LQSYGFRPTYGV | 1971 | 442 |
| HLA-A*68:01 | PTNGVGYQPYR | PTYGVGHQPYR | 1073 | 303 |
| HLA-A*33:03 | PTNGVGYQPYR | PTYGVGHQPYR | 1345 | 417 |
| HLA-A*02:02 | YGFQPTNGV | YGFRPTYGV | 868 | 281 |
| HLA-A*31:01 | GFQPTNGVGYQPYR | GFRPTYGVGHQPYR | 616 | 201 |
| HLA-A*29:02 | PTNGVGYQPY | PTYGVGHQPY | 1211 | 401 |
| HLA-A*31:01 | PTNGVGYQPYR | PTYGVGHQPYR | 1377 | 470 |
| HLA-A*02:06 | YGFQPTNGV | YGFRPTYGV | 174 | 61 |
| HLA-A*68:01 | TNGVGYQPYR | TYGVGHQPYR | 263 | 740 |
| HLA-A*68:02 | YGFQPTNGV | YGFRPTYGV | 73 | 26 |
| HLA-C*12:03 | YGFQPTNGV | YGFRPTYGV | 127 | 355 |
| HLA-A*02:06 | QSYGFQPTNGV | QSYGFRPTYGV | 1121 | 425 |
| HLA-A*68:02 | QSYGFQPTNGV | QSYGFRPTYGV | 122 | 47 |
| HLA-A*30:02 | PTNGVGYQPY | PTYGVGHQPY | 465 | 205 |
| HLA-A*31:01 | NGVGYQPYR | YGVGHQPYR | 438 | 207 |
Y505H
Reference CNGVEGFNCYFPLQSYGFQPTNGVGYQPYRVVVLSFELLHAPATVCGPKKS
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated CNGVAGVNCYFPLQSYGFRPTYGVGHQPYRVVVLSFELLHAPATVCGPKKS
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-C*14:02 | TNGVGYQPY | TYGVGHQPY | 5468 | 16 |
| HLA-B*15:01 | FQPTNGVGY | FRPTYGVGH | 166 | 29043 |
| HLA-B*15:03 | FQPTNGVGY | FRPTYGVGH | 63 | 6390 |
| HLA-A*29:02 | GFQPTNGVGY | GFRPTYGVGH | 188 | 18753 |
| HLA-B*15:01 | LQSYGFQPTNGVGY | LQSYGFRPTYGVGH | 180 | 8335 |
| HLA-C*14:02 | GYQPYRVVV | GHQPYRVVV | 78 | 3539 |
| HLA-B*15:01 | FQPTNGVGYQPY | FRPTYGVGHQPY | 290 | 11634 |
| HLA-C*03:02 | NGVGYQPY | YGVGHQPY | 10463 | 322 |
| HLA-A*24:02 | GYQPYRVVVLSF | GHQPYRVVVLSF | 220 | 6955 |
| HLA-A*23:01 | GYQPYRVVVLSF | GHQPYRVVVLSF | 107 | 3241 |
| HLA-B*15:03 | YGFQPTNGVGY | YGFRPTYGVGH | 368 | 10641 |
| HLA-A*30:02 | GFQPTNGVGY | GFRPTYGVGH | 169 | 4730 |
| HLA-A*30:02 | QSYGFQPTNGVGY | QSYGFRPTYGVGH | 258 | 7114 |
| HLA-B*15:03 | LQSYGFQPTNGVGY | LQSYGFRPTYGVGH | 113 | 2917 |
| HLA-C*14:02 | GYQPYRVVVL | GHQPYRVVVL | 213 | 4430 |
| HLA-C*14:02 | VGYQPYRVVV | VGHQPYRVVV | 413 | 7561 |
| HLA-C*16:01 | NGVGYQPY | YGVGHQPY | 8669 | 488 |
| HLA-C*07:02 | FQPTNGVGYQPY | FRPTYGVGHQPY | 6169 | 356 |
| HLA-A*02:11 | YQPYRVVVL | HQPYRVVVL | 332 | 4009 |
| HLA-A*02:11 | NGVGYQPYRV | YGVGHQPYRV | 5989 | 500 |
| HLA-B*15:03 | NGVGYQPY | YGVGHQPY | 4677 | 418 |
| HLA-A*02:06 | YQPYRVVVL | HQPYRVVVL | 223 | 2206 |
| HLA-C*14:02 | FQPTNGVGYQPY | FRPTYGVGHQPY | 3107 | 366 |
| HLA-A*29:02 | TNGVGYQPY | TYGVGHQPY | 2031 | 243 |
| HLA-A*31:01 | TNGVGYQPYR | TYGVGHQPYR | 811 | 103 |
| HLA-A*02:02 | YQPYRVVVL | HQPYRVVVL | 360 | 2683 |
| HLA-A*33:03 | TNGVGYQPYR | TYGVGHQPYR | 573 | 79 |
| HLA-B*15:03 | FQPTNGVGYQPY | FRPTYGVGHQPY | 183 | 1236 |
| HLA-A*31:01 | QPTNGVGYQPYR | RPTYGVGHQPYR | 3100 | 478 |
| HLA-C*01:02 | YQPYRVVVL | HQPYRVVVL | 41 | 241 |
| HLA-A*02:06 | YQPYRVVVLSFEL | HQPYRVVVLSFEL | 430 | 2495 |
| HLA-C*03:03 | YQPYRVVVL | HQPYRVVVL | 410 | 2128 |
| HLA-C*03:04 | YQPYRVVVL | HQPYRVVVL | 410 | 2128 |
| HLA-C*07:02 | YQPYRVVVL | HQPYRVVVL | 135 | 590 |
| HLA-C*12:03 | YQPYRVVVL | HQPYRVVVL | 132 | 555 |
| HLA-A*68:01 | PTNGVGYQPYR | PTYGVGHQPYR | 1073 | 303 |
| HLA-C*14:02 | YQPYRVVVL | HQPYRVVVL | 102 | 347 |
| HLA-C*16:01 | YQPYRVVVL | HQPYRVVVL | 214 | 726 |
| HLA-A*33:03 | PTNGVGYQPYR | PTYGVGHQPYR | 1345 | 417 |
| HLA-A*31:01 | GFQPTNGVGYQPYR | GFRPTYGVGHQPYR | 616 | 201 |
| HLA-A*29:02 | PTNGVGYQPY | PTYGVGHQPY | 1211 | 401 |
| HLA-C*12:03 | VGYQPYRVV | VGHQPYRVV | 398 | 1196 |
| HLA-A*31:01 | PTNGVGYQPYR | PTYGVGHQPYR | 1377 | 470 |
| HLA-A*68:01 | TNGVGYQPYR | TYGVGHQPYR | 263 | 740 |
| HLA-A*23:01 | YQPYRVVVLSF | HQPYRVVVLSF | 465 | 1280 |
| HLA-A*30:02 | PTNGVGYQPY | PTYGVGHQPY | 465 | 205 |
| HLA-A*31:01 | NGVGYQPYR | YGVGHQPYR | 438 | 207 |
D614G
Reference PCSFGGVSVITPGTNTSNQVAVLYQDVNCTEVPVAIHADQLTPTWRVYSTG
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated PCSFGGVSVITPGTNTSNQVAVLYQGVNCTEVPVAIHADQLTPTWRVYSTG
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-C*05:01 | YQDVNCTEV | YQGVNCTEV | 82 | 8597 |
| HLA-C*08:02 | YQDVNCTEV | YQGVNCTEV | 89 | 8066 |
| HLA-A*02:11 | DVNCTEVPV | GVNCTEVPV | 2140 | 72 |
| HLA-A*02:02 | DVNCTEVPV | GVNCTEVPV | 4099 | 213 |
| HLA-A*68:02 | DVNCTEVPV | GVNCTEVPV | 36 | 659 |
| HLA-A*02:06 | DVNCTEVPV | GVNCTEVPV | 3807 | 208 |
| HLA-A*68:02 | QVAVLYQDV | QVAVLYQGV | 183 | 13 |
| HLA-A*68:02 | NQVAVLYQDV | NQVAVLYQGV | 650 | 52 |
| HLA-A*02:06 | QVAVLYQDV | QVAVLYQGV | 1312 | 148 |
| HLA-A*02:11 | QVAVLYQDV | QVAVLYQGV | 2169 | 268 |
| HLA-A*68:02 | NTSNQVAVLYQDV | NTSNQVAVLYQGV | 1858 | 231 |
| HLA-A*68:02 | TSNQVAVLYQDV | TSNQVAVLYQGV | 3281 | 457 |
| HLA-A*02:02 | QVAVLYQDV | QVAVLYQGV | 1107 | 188 |
| HLA-A*02:01 | YQDVNCTEV | YQGVNCTEV | 75 | 315 |
| HLA-A*02:06 | YQDVNCTEV | YQGVNCTEV | 17 | 57 |
| HLA-A*02:11 | VLYQDVNCT | VLYQGVNCT | 224 | 74 |
| HLA-A*02:11 | YQDVNCTEV | YQGVNCTEV | 31 | 93 |
| HLA-A*02:11 | VLYQDVNCTEVP | VLYQGVNCTEVP | 1000 | 402 |
| HLA-A*02:02 | NQVAVLYQDV | NQVAVLYQGV | 269 | 116 |
| HLA-A*02:11 | AVLYQDVNCTEV | AVLYQGVNCTEV | 500 | 238 |
| HLA-A*02:01 | NQVAVLYQDV | NQVAVLYQGV | 558 | 267 |
| HLA-A*02:06 | NQVAVLYQDV | NQVAVLYQGV | 36 | 18 |
H655Y
Reference TPTWRVYSTGSNVFQTRAGCLIGAEHVNNSYECDIPIGAGICASYQTQTNS
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated TPTWRVYSTGSNVFQTRAGCLIGAEYVNNSYECDIPIGAGICASYQTQTKS
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-B*15:01 | RAGCLIGAEH | RAGCLIGAEY | 7377 | 156 |
| HLA-A*30:02 | RAGCLIGAEH | RAGCLIGAEY | 1881 | 43 |
| HLA-B*15:01 | FQTRAGCLIGAEH | FQTRAGCLIGAEY | 7512 | 178 |
| HLA-B*15:03 | FQTRAGCLIGAEH | FQTRAGCLIGAEY | 5076 | 125 |
| HLA-A*30:02 | AGCLIGAEH | AGCLIGAEY | 5331 | 179 |
| HLA-B*15:03 | RAGCLIGAEH | RAGCLIGAEY | 5254 | 179 |
| HLA-A*30:02 | QTRAGCLIGAEH | QTRAGCLIGAEY | 8425 | 342 |
| HLA-B*15:03 | AGCLIGAEH | AGCLIGAEY | 9025 | 461 |
| HLA-A*02:11 | HVNNSYECDIPI | YVNNSYECDIPI | 1593 | 109 |
| HLA-A*02:01 | HVNNSYECDIPI | YVNNSYECDIPI | 2384 | 226 |
| HLA-A*02:06 | HVNNSYECDIPI | YVNNSYECDIPI | 837 | 92 |
| HLA-A*02:02 | HVNNSYECDIPI | YVNNSYECDIPI | 1466 | 187 |
| HLA-A*02:11 | CLIGAEHV | CLIGAEYV | 983 | 221 |
| HLA-B*15:03 | AEHVNNSY | AEYVNNSY | 986 | 267 |
| HLA-B*18:01 | AEHVNNSY | AEYVNNSY | 902 | 286 |
N679K
Reference EHVNNSYECDIPIGAGICASYQTQTNSPRRARSVASQSIIAYTMSLGAENS
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated EYVNNSYECDIPIGAGICASYQTQTKSHRRARSVASQSIIAYTMSLGAENS
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-A*30:01 | NSPRRARSV | KSHRRARSV | 7195 | 22 |
| HLA-A*68:01 | CASYQTQTN | CASYQTQTK | 28102 | 123 |
| HLA-B*07:02 | TNSPRRARSVA | TKSHRRARSVA | 125 | 19196 |
| HLA-B*07:02 | NSPRRARSVA | KSHRRARSVA | 69 | 5330 |
| HLA-A*11:01 | CASYQTQTN | CASYQTQTK | 32767 | 431 |
| HLA-B*07:02 | TQTNSPRRARSVA | TQTKSHRRARSVA | 270 | 17591 |
| HLA-B*07:02 | QTNSPRRARSVA | QTKSHRRARSVA | 249 | 13621 |
| HLA-B*42:01 | TNSPRRARSVA | TKSHRRARSVA | 437 | 22045 |
| HLA-B*42:01 | NSPRRARSVA | KSHRRARSVA | 217 | 9050 |
| HLA-A*30:01 | NSPRRARSVAS | KSHRRARSVAS | 4546 | 185 |
| HLA-A*30:01 | NSPRRARSVA | KSHRRARSVA | 630 | 27 |
| HLA-C*15:02 | NSPRRARSV | KSHRRARSV | 4094 | 215 |
| HLA-A*68:01 | QTNSPRRAR | QTKSHRRAR | 95 | 1735 |
| HLA-C*14:02 | SYQTQTNSP | SYQTQTKSH | 3691 | 214 |
| HLA-A*68:01 | TQTNSPRRAR | TQTKSHRRAR | 254 | 4244 |
| HLA-A*68:01 | CASYQTQTNSPR | CASYQTQTKSHR | 142 | 713 |
| HLA-C*16:01 | NSPRRARSV | KSHRRARSV | 1247 | 297 |
| HLA-A*11:01 | ASYQTQTNSPR | ASYQTQTKSHR | 346 | 1426 |
| HLA-A*31:01 | QTQTNSPRR | QTQTKSHRR | 355 | 96 |
| HLA-A*33:03 | YQTQTNSPRR | YQTQTKSHRR | 967 | 273 |
| HLA-A*33:03 | YQTQTNSPRRAR | YQTQTKSHRRAR | 306 | 1077 |
| HLA-A*31:01 | YQTQTNSPRR | YQTQTKSHRR | 965 | 276 |
| HLA-A*33:03 | TQTNSPRRAR | TQTKSHRRAR | 195 | 659 |
| HLA-A*33:03 | QTQTNSPRR | QTQTKSHRR | 445 | 143 |
| HLA-A*31:01 | ASYQTQTNSPRR | ASYQTQTKSHRR | 433 | 144 |
| HLA-A*33:03 | QTNSPRRAR | QTKSHRRAR | 48 | 144 |
| HLA-A*30:01 | QTNSPRRAR | QTKSHRRAR | 1146 | 463 |
| HLA-A*31:01 | YQTQTNSPRRAR | YQTQTKSHRRAR | 317 | 771 |
| HLA-A*31:01 | SYQTQTNSPR | SYQTQTKSHR | 62 | 147 |
| HLA-A*33:03 | SYQTQTNSPR | SYQTQTKSHR | 130 | 304 |
| HLA-A*31:01 | TQTNSPRRAR | TQTKSHRRAR | 144 | 336 |
| HLA-A*31:01 | QTNSPRRAR | QTKSHRRAR | 34 | 79 |
| HLA-A*31:01 | ASYQTQTNSPR | ASYQTQTKSHR | 139 | 315 |
| HLA-A*31:01 | YQTQTNSPR | YQTQTKSHR | 441 | 982 |
| HLA-A*31:01 | SYQTQTNSPRRAR | SYQTQTKSHRRAR | 193 | 427 |
| HLA-A*33:03 | SYQTQTNSPRRAR | SYQTQTKSHRRAR | 448 | 972 |
P681H
Reference VNNSYECDIPIGAGICASYQTQTNSPRRARSVASQSIIAYTMSLGAENSVA
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated VNNSYECDIPIGAGICASYQTQTKSHRRARSVASQSIIAYTMSLGAENSVA
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-B*07:02 | SPRRARSVA | SHRRARSVA | 4 | 4721 |
| HLA-B*42:01 | SPRRARSVA | SHRRARSVA | 8 | 6169 |
| HLA-B*07:02 | SPRRARSVASQSI | SHRRARSVASQSI | 17 | 11433 |
| HLA-B*07:02 | SPRRARSV | SHRRARSV | 24 | 12728 |
| HLA-B*07:02 | SPRRARSVAS | SHRRARSVAS | 37 | 14758 |
| HLA-A*30:01 | NSPRRARSV | KSHRRARSV | 7195 | 22 |
| HLA-B*07:02 | SPRRARSVASQSII | SHRRARSVASQSII | 55 | 17736 |
| HLA-B*42:01 | SPRRARSV | SHRRARSV | 38 | 11622 |
| HLA-B*42:01 | SPRRARSVASQSI | SHRRARSVASQSI | 48 | 14418 |
| HLA-B*07:02 | TNSPRRARSVA | TKSHRRARSVA | 125 | 19196 |
| HLA-B*42:01 | SPRRARSVAS | SHRRARSVAS | 158 | 17461 |
| HLA-B*07:02 | NSPRRARSVA | KSHRRARSVA | 69 | 5330 |
| HLA-B*42:01 | SPRRARSVASQSII | SHRRARSVASQSII | 321 | 21521 |
| HLA-B*07:02 | TQTNSPRRARSVA | TQTKSHRRARSVA | 270 | 17591 |
| HLA-B*07:02 | QTNSPRRARSVA | QTKSHRRARSVA | 249 | 13621 |
| HLA-B*42:01 | TNSPRRARSVA | TKSHRRARSVA | 437 | 22045 |
| HLA-B*42:01 | NSPRRARSVA | KSHRRARSVA | 217 | 9050 |
| HLA-A*30:01 | NSPRRARSVAS | KSHRRARSVAS | 4546 | 185 |
| HLA-A*30:01 | NSPRRARSVA | KSHRRARSVA | 630 | 27 |
| HLA-C*15:02 | NSPRRARSV | KSHRRARSV | 4094 | 215 |
| HLA-A*68:01 | QTNSPRRAR | QTKSHRRAR | 95 | 1735 |
| HLA-C*14:02 | SYQTQTNSP | SYQTQTKSH | 3691 | 214 |
| HLA-A*68:01 | TQTNSPRRAR | TQTKSHRRAR | 254 | 4244 |
| HLA-B*08:01 | SPRRARSVA | SHRRARSVA | 372 | 3831 |
| HLA-B*08:01 | SPRRARSV | SHRRARSV | 428 | 2448 |
| HLA-A*68:01 | CASYQTQTNSPR | CASYQTQTKSHR | 142 | 713 |
| HLA-C*16:01 | NSPRRARSV | KSHRRARSV | 1247 | 297 |
| HLA-A*11:01 | ASYQTQTNSPR | ASYQTQTKSHR | 346 | 1426 |
| HLA-A*31:01 | QTQTNSPRR | QTQTKSHRR | 355 | 96 |
| HLA-A*33:03 | YQTQTNSPRR | YQTQTKSHRR | 967 | 273 |
| HLA-A*33:03 | YQTQTNSPRRAR | YQTQTKSHRRAR | 306 | 1077 |
| HLA-A*31:01 | YQTQTNSPRR | YQTQTKSHRR | 965 | 276 |
| HLA-A*33:03 | TQTNSPRRAR | TQTKSHRRAR | 195 | 659 |
| HLA-A*33:03 | QTQTNSPRR | QTQTKSHRR | 445 | 143 |
| HLA-A*31:01 | ASYQTQTNSPRR | ASYQTQTKSHRR | 433 | 144 |
| HLA-A*33:03 | QTNSPRRAR | QTKSHRRAR | 48 | 144 |
| HLA-A*30:01 | QTNSPRRAR | QTKSHRRAR | 1146 | 463 |
| HLA-A*31:01 | YQTQTNSPRRAR | YQTQTKSHRRAR | 317 | 771 |
| HLA-A*31:01 | SYQTQTNSPR | SYQTQTKSHR | 62 | 147 |
| HLA-A*33:03 | SYQTQTNSPR | SYQTQTKSHR | 130 | 304 |
| HLA-A*31:01 | TQTNSPRRAR | TQTKSHRRAR | 144 | 336 |
| HLA-A*31:01 | QTNSPRRAR | QTKSHRRAR | 34 | 79 |
| HLA-A*31:01 | ASYQTQTNSPR | ASYQTQTKSHR | 139 | 315 |
| HLA-A*31:01 | YQTQTNSPR | YQTQTKSHR | 441 | 982 |
| HLA-A*31:01 | SYQTQTNSPRRAR | SYQTQTKSHRRAR | 193 | 427 |
| HLA-A*33:03 | SYQTQTNSPRRAR | SYQTQTKSHRRAR | 448 | 972 |
N764K
Reference TMYICGDSTECSNLLLQYGSFCTQLNRALTGIAVEQDKNTQEVFAQVKQIY
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated TMYICGDSTECSNLLLQYGSFCTQLKRALTGIAVEQDKNTQEVFAQVKQIY
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-A*03:02 | LLQYGSFCTQLN | LLQYGSFCTQLK | 29153 | 102 |
| HLA-A*11:01 | LQYGSFCTQLN | LQYGSFCTQLK | 32767 | 191 |
| HLA-A*03:01 | LQYGSFCTQLN | LQYGSFCTQLK | 28204 | 214 |
| HLA-A*68:01 | YGSFCTQLN | YGSFCTQLK | 23773 | 187 |
| HLA-A*03:02 | LQYGSFCTQLN | LQYGSFCTQLK | 31227 | 331 |
| HLA-A*03:02 | LLLQYGSFCTQLN | LLLQYGSFCTQLK | 32767 | 377 |
| HLA-A*11:01 | YGSFCTQLN | YGSFCTQLK | 32767 | 386 |
| HLA-A*03:01 | LLQYGSFCTQLN | LLQYGSFCTQLK | 29424 | 350 |
| HLA-A*03:01 | LLLQYGSFCTQLN | LLLQYGSFCTQLK | 32355 | 410 |
| HLA-A*02:11 | QLNRALTGI | QLKRALTGI | 27 | 440 |
| HLA-A*02:11 | FCTQLNRALTGI | FCTQLKRALTGI | 63 | 898 |
| HLA-B*08:01 | CTQLNRAL | CTQLKRAL | 3325 | 279 |
| HLA-A*02:01 | FCTQLNRALTGI | FCTQLKRALTGI | 493 | 4878 |
| HLA-A*02:02 | FCTQLNRALTGI | FCTQLKRALTGI | 62 | 503 |
| HLA-A*02:02 | QLNRALTGI | QLKRALTGI | 45 | 307 |
| HLA-C*03:03 | FCTQLNRAL | FCTQLKRAL | 138 | 752 |
| HLA-C*03:04 | FCTQLNRAL | FCTQLKRAL | 138 | 752 |
| HLA-A*02:02 | TQLNRALTGI | TQLKRALTGI | 180 | 882 |
| HLA-A*02:11 | TQLNRALTGI | TQLKRALTGI | 267 | 1284 |
| HLA-B*27:05 | NRALTGIAV | KRALTGIAV | 1251 | 276 |
| HLA-A*02:11 | QLNRALTGIAV | QLKRALTGIAV | 259 | 1123 |
| HLA-C*03:02 | FCTQLNRAL | FCTQLKRAL | 361 | 1438 |
| HLA-C*14:02 | SFCTQLNRAL | SFCTQLKRAL | 234 | 511 |
| HLA-A*02:06 | TQLNRALTGI | TQLKRALTGI | 54 | 113 |
D796Y
Reference AVEQDKNTQEVFAQVKQIYKTPPIKDFGGFNFSQILPDPSKPSKRSFIEDL
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated AVEQDKNTQEVFAQVKQIYKTPPIKYFGGFNFSQILPDPSKPSKRSFIEDL
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-A*23:01 | KDFGGFNF | KYFGGFNF | 15419 | 23 |
| HLA-A*23:01 | KDFGGFNFSQI | KYFGGFNFSQI | 15932 | 37 |
| HLA-A*24:02 | KDFGGFNF | KYFGGFNF | 21008 | 55 |
| HLA-C*14:02 | IYKTPPIKD | IYKTPPIKY | 16538 | 55 |
| HLA-A*24:02 | KDFGGFNFSQI | KYFGGFNFSQI | 20251 | 68 |
| HLA-B*15:03 | KQIYKTPPIKD | KQIYKTPPIKY | 7773 | 29 |
| HLA-A*23:01 | KDFGGFNFSQIL | KYFGGFNFSQIL | 15831 | 63 |
| HLA-C*14:02 | KDFGGFNF | KYFGGFNF | 22081 | 90 |
| HLA-A*24:02 | KDFGGFNFSQIL | KYFGGFNFSQIL | 21565 | 126 |
| HLA-B*15:01 | KQIYKTPPIKD | KQIYKTPPIKY | 22193 | 141 |
| HLA-C*14:02 | KDFGGFNFSQIL | KYFGGFNFSQIL | 14873 | 165 |
| HLA-A*30:01 | KDFGGFNFS | KYFGGFNFS | 9496 | 156 |
| HLA-C*14:02 | DFGGFNFSQIL | YFGGFNFSQIL | 6960 | 139 |
| HLA-A*30:02 | KQIYKTPPIKD | KQIYKTPPIKY | 15917 | 386 |
| HLA-B*15:03 | VKQIYKTPPIKD | VKQIYKTPPIKY | 16977 | 446 |
| HLA-B*15:03 | IKDFGGFNF | IKYFGGFNF | 97 | 4 |
| HLA-B*15:03 | PIKDFGGFNF | PIKYFGGFNF | 7546 | 321 |
| HLA-B*15:03 | TPPIKDFGGFNF | TPPIKYFGGFNF | 10071 | 484 |
| HLA-B*15:03 | KTPPIKDFGGFNF | KTPPIKYFGGFNF | 6427 | 331 |
| HLA-A*32:01 | KTPPIKDFGGFNF | KTPPIKYFGGFNF | 6031 | 381 |
| HLA-A*23:01 | DFGGFNFSQI | YFGGFNFSQI | 5424 | 461 |
| HLA-B*15:03 | YKTPPIKDFGGFNF | YKTPPIKYFGGFNF | 2588 | 394 |
| HLA-C*06:02 | YKTPPIKDF | YKTPPIKYF | 2488 | 460 |
| HLA-C*07:02 | YKTPPIKDF | YKTPPIKYF | 1804 | 378 |
| HLA-A*23:01 | IYKTPPIKDF | IYKTPPIKYF | 273 | 98 |
| HLA-A*24:02 | IYKTPPIKDF | IYKTPPIKYF | 375 | 147 |
| HLA-C*14:02 | IYKTPPIKDF | IYKTPPIKYF | 341 | 158 |
| HLA-B*15:03 | YKTPPIKDF | YKTPPIKYF | 92 | 43 |
Q954H
Reference SAIGKIQDSLSSTASALGKLQDVVNQNAQALNTLVKQLSSNFGAISSVLND
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated SAIGKIQDSLSSTASALGKLQDVVNHNAQALNTLVKQLSSKFGAISSVLND
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-B*38:01 | NQNAQALNTL | NHNAQALNTL | 2805 | 97 |
| HLA-A*68:02 | DVVNQNAQA | DVVNHNAQA | 703 | 344 |
N969K
Reference ALGKLQDVVNQNAQALNTLVKQLSSNFGAISSVLNDILSRLDKVEAEVQID
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated ALGKLQDVVNHNAQALNTLVKQLSSKFGAISSVLNDILSRLDKVEAEVQID
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-A*03:01 | TLVKQLSSN | TLVKQLSSK | 29207 | 56 |
| HLA-A*68:01 | NTLVKQLSSN | NTLVKQLSSK | 28063 | 86 |
| HLA-A*11:01 | TLVKQLSSN | TLVKQLSSK | 32767 | 125 |
| HLA-A*03:02 | TLVKQLSSN | TLVKQLSSK | 31915 | 131 |
| HLA-A*11:01 | NTLVKQLSSN | NTLVKQLSSK | 32767 | 147 |
| HLA-A*03:01 | ALNTLVKQLSSN | ALNTLVKQLSSK | 32767 | 154 |
| HLA-A*03:02 | ALNTLVKQLSSN | ALNTLVKQLSSK | 32767 | 304 |
| HLA-A*03:01 | NTLVKQLSSN | NTLVKQLSSK | 32767 | 482 |
| HLA-A*68:02 | SNFGAISSV | SKFGAISSV | 117 | 5329 |
| HLA-B*15:03 | SNFGAISSVL | SKFGAISSVL | 688 | 25 |
| HLA-B*08:01 | QLSSNFGAI | QLSSKFGAI | 4755 | 265 |
| HLA-C*15:02 | SNFGAISSV | SKFGAISSV | 374 | 5700 |
| HLA-A*30:01 | SSNFGAISS | SSKFGAISS | 1320 | 88 |
| HLA-B*15:03 | SNFGAISSV | SKFGAISSV | 727 | 49 |
| HLA-A*30:01 | SSNFGAISSV | SSKFGAISSV | 587 | 71 |
| HLA-A*02:06 | KQLSSNFGA | KQLSSKFGA | 62 | 269 |
| HLA-A*02:01 | KQLSSNFGAI | KQLSSKFGAI | 407 | 1567 |
| HLA-A*02:11 | KQLSSNFGA | KQLSSKFGA | 228 | 807 |
| HLA-A*02:06 | SNFGAISSV | SKFGAISSV | 361 | 1100 |
| HLA-C*06:02 | SNFGAISSV | SKFGAISSV | 1423 | 485 |
| HLA-A*02:11 | KQLSSNFGAI | KQLSSKFGAI | 152 | 428 |
| HLA-A*02:06 | KQLSSNFGAI | KQLSSKFGAI | 58 | 159 |
| HLA-A*02:02 | KQLSSNFGAI | KQLSSKFGAI | 261 | 668 |
| HLA-B*15:03 | KQLSSNFGA | KQLSSKFGA | 497 | 1076 |
N protein
P13L
Reference MSDNGPQNQRNAPRITFGGPSDSTGSNQNGERSGARSK
|||||||||||| |||||||||||||||||||||||||
Mutated MSDNGPQNQRNALRITFGGPSDSTGSNQNG‑‑‑GARSK
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-B*42:01 | MSDNGPQNQRNAP | MSDNGPQNQRNAL | 11146 | 27 |
| HLA-B*07:02 | GPQNQRNAP | GPQNQRNAL | 2464 | 6 |
| HLA-B*42:01 | GPQNQRNAP | GPQNQRNAL | 3035 | 8 |
| HLA-B*07:02 | SDNGPQNQRNAP | SDNGPQNQRNAL | 11353 | 32 |
| HLA-B*42:01 | SDNGPQNQRNAP | SDNGPQNQRNAL | 13042 | 39 |
| HLA-B*42:01 | NGPQNQRNAP | NGPQNQRNAL | 12875 | 41 |
| HLA-B*07:02 | MSDNGPQNQRNAP | MSDNGPQNQRNAL | 13138 | 58 |
| HLA-B*07:02 | NGPQNQRNAP | NGPQNQRNAL | 14898 | 72 |
| HLA-B*07:02 | APRITFGGPS | ALRITFGGPS | 191 | 17534 |
| HLA-B*42:01 | PQNQRNAP | PQNQRNAL | 30142 | 436 |
| HLA-B*07:02 | GPQNQRNAPR | GPQNQRNALR | 13964 | 345 |
| HLA-B*27:05 | QRNAPRITF | QRNALRITF | 509 | 83 |
| HLA-B*27:05 | NQRNAPRITF | NQRNALRITF | 1716 | 350 |
| HLA-B*27:05 | GPQNQRNAPRITF | GPQNQRNALRITF | 2076 | 442 |
| HLA-B*15:01 | NQRNAPRITF | NQRNALRITF | 96 | 40 |
ERS 31-33 deletion
Reference PQNQRNAPRITFGGPSDSTGSNQNGERSGARSKQRRPQGLPNNTASWFTALTQ
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated PQNQRNALRITFGGPSDSTGSNQNG‑‑‑GARSKQRRPQGLPNNTASWFTALTQ
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-A*31:01 | RSGARSKQR | GARSKQR | 77 | - |
| HLA-A*68:01 | STGSNQNGER | - | 137 | - |
| HLA-A*31:01 | RSGARSKQRR | - | 254 | - |
P151S
Reference NKDGIIWVATEGALNTPKDHIGTRNPANNAAIVLQLPQGTTLPKGFYAEGS
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated NKDGIIWVATEGALNTPKDHIGTRNSANNAAIVLQLPQGTTLPKGFYAEGS
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-C*03:03 | PANNAAIVL | SANNAAIVL | 3749 | 9 |
| HLA-C*03:04 | PANNAAIVL | SANNAAIVL | 3749 | 9 |
| HLA-C*16:01 | PANNAAIVL | SANNAAIVL | 1730 | 8 |
| HLA-C*12:03 | PANNAAIVL | SANNAAIVL | 5617 | 27 |
| HLA-C*03:02 | PANNAAIVL | SANNAAIVL | 5558 | 30 |
| HLA-B*07:02 | RNPANNAAIV | RNSANNAAIV | 199 | 19874 |
| HLA-C*12:02 | PANNAAIVL | SANNAAIVL | 10293 | 123 |
| HLA-C*17:01 | PANNAAIVL | SANNAAIVL | 5437 | 65 |
| HLA-B*15:03 | PANNAAIVL | SANNAAIVL | 10239 | 129 |
| HLA-B*42:01 | NPANNAAIVL | NSANNAAIVL | 144 | 9452 |
| HLA-B*42:01 | RNPANNAAIV | RNSANNAAIV | 340 | 20370 |
| HLA-C*15:02 | PANNAAIVL | SANNAAIVL | 10150 | 225 |
| HLA-B*42:01 | NPANNAAIV | NSANNAAIV | 364 | 16078 |
| HLA-B*07:02 | NPANNAAIVL | NSANNAAIVL | 346 | 15227 |
| HLA-A*68:02 | NPANNAAIV | NSANNAAIV | 3031 | 72 |
| HLA-C*08:01 | PANNAAIVL | SANNAAIVL | 12500 | 326 |
| HLA-C*02:02 | PANNAAIVL | SANNAAIVL | 13200 | 452 |
| HLA-C*02:10 | PANNAAIVL | SANNAAIVL | 13200 | 452 |
| HLA-C*14:02 | PANNAAIVL | SANNAAIVL | 10601 | 476 |
| HLA-C*12:03 | NPANNAAIV | NSANNAAIV | 8213 | 461 |
| HLA-C*16:01 | NPANNAAIV | NSANNAAIV | 7224 | 478 |
203-204 RG->KR
Reference GGSQASSRSSSRSRNSSRNSTPGSSRGTSPARMAGNGGDAALALLLLDRLNQ
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated GGSQASSRSSSRSRNSSRNSTPGSSKRTSPARMAGNGGDAALALLLLDRLNQ
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-A*68:01 | NSTPGSSRG | NSTPGSSKR | 32767 | 34 |
| HLA-A*33:03 | NSTPGSSRG | NSTPGSSKR | 32767 | 300 |
| HLA-B*15:03 | SRGTSPARM | SKRTSPARM | 2669 | 106 |
| HLA-A*31:01 | RNSTPGSSRG | RNSTPGSSKR | 7013 | 340 |
| HLA-A*30:01 | SSRNSTPGSSR | SSRNSTPGSSK | 356 | 20 |
| HLA-A*30:01 | GTSPARMAG | RTSPARMAG | 493 | 30 |
| HLA-A*31:01 | SSRNSTPGSSRG | SSRNSTPGSSKR | 3757 | 231 |
| HLA-A*30:01 | GTSPARMAGNGG | RTSPARMAGNGG | 6051 | 411 |
| HLA-A*31:01 | SSRNSTPGSSR | SSRNSTPGSSK | 127 | 1853 |
| HLA-A*30:01 | NSSRNSTPGSSR | NSSRNSTPGSSK | 4255 | 353 |
| HLA-A*30:01 | RNSTPGSSR | RNSTPGSSK | 2120 | 188 |
| HLA-A*31:01 | RNSTPGSSR | RNSTPGSSK | 123 | 1335 |
| HLA-A*03:01 | SSRNSTPGSSR | SSRNSTPGSSK | 4753 | 442 |
| HLA-A*11:01 | SSRNSTPGSSR | SSRNSTPGSSK | 2231 | 263 |
| HLA-A*68:01 | NSSRNSTPGSSR | NSSRNSTPGSSK | 270 | 1385 |
| HLA-A*30:01 | GSSRGTSPA | GSSKRTSPA | 555 | 145 |
| HLA-A*30:01 | SSRNSTPGSSRG | SSRNSTPGSSKR | 1625 | 469 |
| HLA-A*30:01 | SSRGTSPARMAG | SSKRTSPARMAG | 449 | 1031 |
S413R
Reference KQQTVTLLPAADLDDFSKQLQQSMSSADSTQA
||||||||||||||||||||||||| ||||||
Mutated KQQTVTLLPAADLDDFSKQLQQSMSRADSTQA
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-A*31:01 | KQLQQSMSS | KQLQQSMSR | 20016 | 55 |
| HLA-A*30:01 | MSSADSTQA | MSRADSTQA | 2953 | 37 |
| HLA-A*33:03 | DFSKQLQQSMSS | DFSKQLQQSMSR | 32767 | 494 |
| HLA-B*15:03 | KQLQQSMSS | KQLQQSMSR | 236 | 5105 |
| HLA-A*30:01 | SMSSADSTQA | SMSRADSTQA | 6453 | 416 |
| HLA-A*02:02 | FSKQLQQSMSSA | FSKQLQQSMSRA | 345 | 1138 |
| HLA-A*02:11 | FSKQLQQSMSSA | FSKQLQQSMSRA | 353 | 1118 |
| HLA-A*02:11 | QLQQSMSSA | QLQQSMSRA | 436 | 1175 |
| HLA-A*02:02 | SMSSADSTQA | SMSRADSTQA | 355 | 725 |
M protein
Q19E
Reference MADSNGTITVEELKKLLEQWNLVIGFLFLTWICLLQFAYANRNR
|||||||||||||||||| |||||||||||||||||||||||||
Mutated MADSNGTITVEELKKLLEEWNLVIGFLFLTWICLLQFAYANRNR
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-B*44:03 | EQWNLVIGF | EEWNLVIGF | 1268 | 15 |
| HLA-B*44:02 | EQWNLVIGF | EEWNLVIGF | 2396 | 34 |
| HLA-B*44:03 | EQWNLVIGFLF | EEWNLVIGFLF | 5200 | 85 |
| HLA-B*44:03 | EQWNLVIGFL | EEWNLVIGFL | 8038 | 135 |
| HLA-B*44:03 | EQWNLVIGFLFLTW | EEWNLVIGFLFLTW | 5966 | 126 |
| HLA-B*44:02 | EQWNLVIGFLF | EEWNLVIGFLF | 6607 | 162 |
| HLA-B*44:02 | EQWNLVIGFL | EEWNLVIGFL | 9054 | 237 |
| HLA-B*44:02 | EQWNLVIGFLFLTW | EEWNLVIGFLFLTW | 5922 | 170 |
| HLA-B*15:01 | LEQWNLVIGF | LEEWNLVIGF | 256 | 8100 |
| HLA-B*18:01 | EQWNLVIGF | EEWNLVIGF | 482 | 16 |
| HLA-A*02:06 | EQWNLVIGFL | EEWNLVIGFL | 418 | 10916 |
| HLA-B*15:03 | EQWNLVIGF | EEWNLVIGF | 101 | 2397 |
| HLA-B*18:01 | EQWNLVIGFLF | EEWNLVIGFLF | 3471 | 190 |
| HLA-B*40:02 | EQWNLVIGFL | EEWNLVIGFL | 3775 | 245 |
| HLA-B*15:03 | LEQWNLVIGF | LEEWNLVIGF | 88 | 1227 |
| HLA-B*40:02 | EQWNLVIGF | EEWNLVIGF | 1765 | 242 |
| HLA-B*15:03 | KLLEQWNLVIGF | KLLEEWNLVIGF | 365 | 2285 |
| HLA-A*02:11 | LLEQWNLVI | LLEEWNLVI | 142 | 24 |
| HLA-A*02:11 | LLEQWNLVIGFL | LLEEWNLVIGFL | 1120 | 210 |
| HLA-A*02:01 | LLEQWNLVI | LLEEWNLVI | 644 | 131 |
| HLA-A*29:02 | QWNLVIGFLF | EWNLVIGFLF | 159 | 780 |
| HLA-A*24:02 | QWNLVIGFLF | EWNLVIGFLF | 53 | 246 |
| HLA-A*23:01 | QWNLVIGFLF | EWNLVIGFLF | 18 | 79 |
| HLA-A*02:02 | LLEQWNLVI | LLEEWNLVI | 781 | 187 |
| HLA-A*02:06 | LLEQWNLVI | LLEEWNLVI | 1869 | 487 |
| HLA-B*40:02 | LEQWNLVI | LEEWNLVI | 367 | 1301 |
| HLA-A*02:02 | LLEQWNLVIGFL | LLEEWNLVIGFL | 1047 | 329 |
A63T
Reference AYANRNRFLYIIKLIFLWLLWPVTLACFVLAAVYRINWITGGIAIAMACLV
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated AYANRNRFLYIIKLIFLWLLWPVTLTCFVLAAVYRINWITGGIAIAMACLV
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-B*35:01 | LACFVLAAVY | LTCFVLAAVY | 121 | 1943 |
| HLA-A*68:01 | ACFVLAAVYR | TCFVLAAVYR | 694 | 80 |
| HLA-B*35:01 | ACFVLAAVY | TCFVLAAVY | 2289 | 331 |
| HLA-C*03:02 | LACFVLAAVY | LTCFVLAAVY | 145 | 935 |
| HLA-C*16:01 | LACFVLAAV | LTCFVLAAV | 346 | 1994 |
| HLA-A*02:01 | WLLWPVTLA | WLLWPVTLT | 33 | 190 |
| HLA-A*02:02 | WLLWPVTLA | WLLWPVTLT | 32 | 180 |
| HLA-A*02:11 | LWLLWPVTLA | LWLLWPVTLT | 66 | 363 |
| HLA-A*02:11 | IFLWLLWPVTLA | IFLWLLWPVTLT | 190 | 1034 |
| HLA-A*02:11 | LLWPVTLA | LLWPVTLT | 242 | 1313 |
| HLA-A*33:03 | ACFVLAAVYR | TCFVLAAVYR | 252 | 47 |
| HLA-A*02:02 | LWLLWPVTLA | LWLLWPVTLT | 304 | 1623 |
| HLA-C*12:03 | LACFVLAAV | LTCFVLAAV | 214 | 1116 |
| HLA-A*02:06 | WLLWPVTLA | WLLWPVTLT | 60 | 302 |
| HLA-A*02:01 | LWLLWPVTLA | LWLLWPVTLT | 228 | 1142 |
| HLA-A*02:02 | TLACFVLAAVY | TLTCFVLAAVY | 337 | 1467 |
| HLA-A*02:02 | VTLACFVLAAV | VTLTCFVLAAV | 160 | 678 |
| HLA-A*68:02 | LACFVLAAV | LTCFVLAAV | 44 | 11 |
| HLA-A*02:02 | PVTLACFVLAAV | PVTLTCFVLAAV | 479 | 1907 |
| HLA-A*02:11 | LIFLWLLWPVTLA | LIFLWLLWPVTLT | 406 | 1602 |
| HLA-A*02:02 | TLACFVLAAVYRI | TLTCFVLAAVYRI | 293 | 1148 |
| HLA-A*02:11 | LACFVLAAV | LTCFVLAAV | 122 | 32 |
| HLA-A*02:11 | WLLWPVTLA | WLLWPVTLT | 6 | 22 |
| HLA-B*15:03 | LACFVLAAVY | LTCFVLAAVY | 81 | 284 |
| HLA-A*68:02 | TLACFVLAA | TLTCFVLAA | 395 | 1376 |
| HLA-A*02:02 | FLWLLWPVTLACFV | FLWLLWPVTLTCFV | 485 | 146 |
| HLA-A*68:01 | LACFVLAAVY | LTCFVLAAVY | 1184 | 359 |
| HLA-A*02:02 | TLACFVLAA | TLTCFVLAA | 13 | 41 |
| HLA-A*02:11 | TLACFVLAAVY | TLTCFVLAAVY | 310 | 929 |
| HLA-A*02:02 | FLWLLWPVTLA | FLWLLWPVTLT | 291 | 867 |
| HLA-A*02:11 | KLIFLWLLWPVTLA | KLIFLWLLWPVTLT | 290 | 859 |
| HLA-A*29:02 | LACFVLAAVY | LTCFVLAAVY | 232 | 81 |
| HLA-A*02:02 | VTLACFVLAA | VTLTCFVLAA | 100 | 285 |
| HLA-A*02:06 | TLACFVLAAV | TLTCFVLAAV | 16 | 45 |
| HLA-A*68:02 | VTLACFVLAA | VTLTCFVLAA | 481 | 1321 |
| HLA-B*15:01 | TLACFVLAAVY | TLTCFVLAAVY | 414 | 1136 |
| HLA-A*23:01 | LWPVTLACF | LWPVTLTCF | 57 | 155 |
| HLA-A*24:02 | LWPVTLACF | LWPVTLTCF | 49 | 133 |
| HLA-A*02:11 | FLWLLWPVTLA | FLWLLWPVTLT | 25 | 67 |
| HLA-A*02:06 | TLACFVLAA | TLTCFVLAA | 40 | 107 |
| HLA-A*02:01 | LACFVLAAV | LTCFVLAAV | 471 | 179 |
| HLA-A*02:02 | TLACFVLAAV | TLTCFVLAAV | 5 | 13 |
| HLA-A*02:11 | VTLACFVLAA | VTLTCFVLAA | 97 | 245 |
| HLA-A*02:01 | FLWLLWPVTLA | FLWLLWPVTLT | 107 | 269 |
| HLA-A*02:06 | FLWLLWPVTLA | FLWLLWPVTLT | 495 | 1240 |
| HLA-A*02:11 | TLACFVLAA | TLTCFVLAA | 8 | 20 |
| HLA-A*02:11 | VTLACFVLAAV | VTLTCFVLAAV | 128 | 318 |
| HLA-A*02:01 | TLACFVLAAV | TLTCFVLAAV | 11 | 27 |
| HLA-A*02:02 | LACFVLAAV | LTCFVLAAV | 198 | 81 |
| HLA-A*02:01 | TLACFVLAA | TLTCFVLAA | 25 | 61 |
| HLA-A*02:01 | VTLACFVLAAV | VTLTCFVLAAV | 436 | 1029 |
| HLA-B*07:02 | WPVTLACFVL | WPVTLTCFVL | 362 | 155 |
| HLA-A*02:01 | FLWLLWPVTLACFV | FLWLLWPVTLTCFV | 329 | 142 |
| HLA-A*02:11 | TLACFVLAAV | TLTCFVLAAV | 4 | 9 |
| HLA-A*30:02 | LACFVLAAVY | LTCFVLAAVY | 218 | 98 |
| HLA-A*02:06 | LACFVLAAV | LTCFVLAAV | 51 | 23 |
| HLA-A*03:02 | TLACFVLAAVYR | TLTCFVLAAVYR | 480 | 1047 |
| HLA-A*68:02 | TLACFVLAAV | TLTCFVLAAV | 26 | 12 |
| HLA-B*42:01 | WPVTLACFV | WPVTLTCFV | 209 | 102 |
| HLA-A*32:01 | LLWPVTLACF | LLWPVTLTCF | 427 | 212 |
E protein

T9I
Reference MYSFVSEETGTLIVNSVLLFLAFVVFLLVTLAIL
|||||||| |||||||||||||||||||||||||
Mutated MYSFVSEEIGTLIVNSVLLFLAFVVFLLVTLAIL
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-A*24:02 | MYSFVSEET | MYSFVSEEI | 5532 | 18 |
| HLA-A*23:01 | MYSFVSEET | MYSFVSEEI | 5604 | 31 |
| HLA-C*16:01 | YSFVSEET | YSFVSEEI | 7157 | 226 |
| HLA-C*14:02 | MYSFVSEET | MYSFVSEEI | 901 | 30 |
| HLA-C*12:03 | YSFVSEET | YSFVSEEI | 10273 | 457 |
| HLA-A*68:02 | MYSFVSEET | MYSFVSEEI | 3879 | 472 |
| HLA-A*68:02 | ETGTLIVNSV | EIGTLIVNSV | 13 | 91 |
| HLA-A*68:02 | FVSEETGTLI | FVSEEIGTLI | 247 | 60 |
| HLA-A*68:02 | TGTLIVNSV | IGTLIVNSV | 491 | 1976 |
| HLA-A*02:11 | FVSEETGTLI | FVSEEIGTLI | 307 | 89 |
| HLA-B*40:01 | EETGTLIVNSVL | EEIGTLIVNSVL | 1578 | 499 |
| HLA-A*02:01 | FVSEETGTL | FVSEEIGTL | 575 | 186 |
| HLA-A*02:11 | FVSEETGTL | FVSEEIGTL | 97 | 32 |
| HLA-A*02:06 | FVSEETGTLI | FVSEEIGTLI | 423 | 144 |
| HLA-A*02:06 | FVSEETGTL | FVSEEIGTL | 120 | 41 |
| HLA-A*02:01 | FVSEETGTLI | FVSEEIGTLI | 1293 | 442 |
| HLA-A*02:06 | FVSEETGTLIV | FVSEEIGTLIV | 670 | 266 |
| HLA-C*03:03 | YSFVSEETGTL | YSFVSEEIGTL | 293 | 711 |
| HLA-C*03:04 | YSFVSEETGTL | YSFVSEEIGTL | 293 | 711 |
| HLA-B*44:03 | EETGTLIVNSVLLF | EEIGTLIVNSVLLF | 877 | 375 |
| HLA-A*02:11 | FVSEETGTLIV | FVSEEIGTLIV | 538 | 251 |
| HLA-A*02:02 | FVSEETGTLI | FVSEEIGTLI | 85 | 40 |
| HLA-A*68:02 | YSFVSEETGTLI | YSFVSEEIGTLI | 1008 | 485 |
NS3 protein
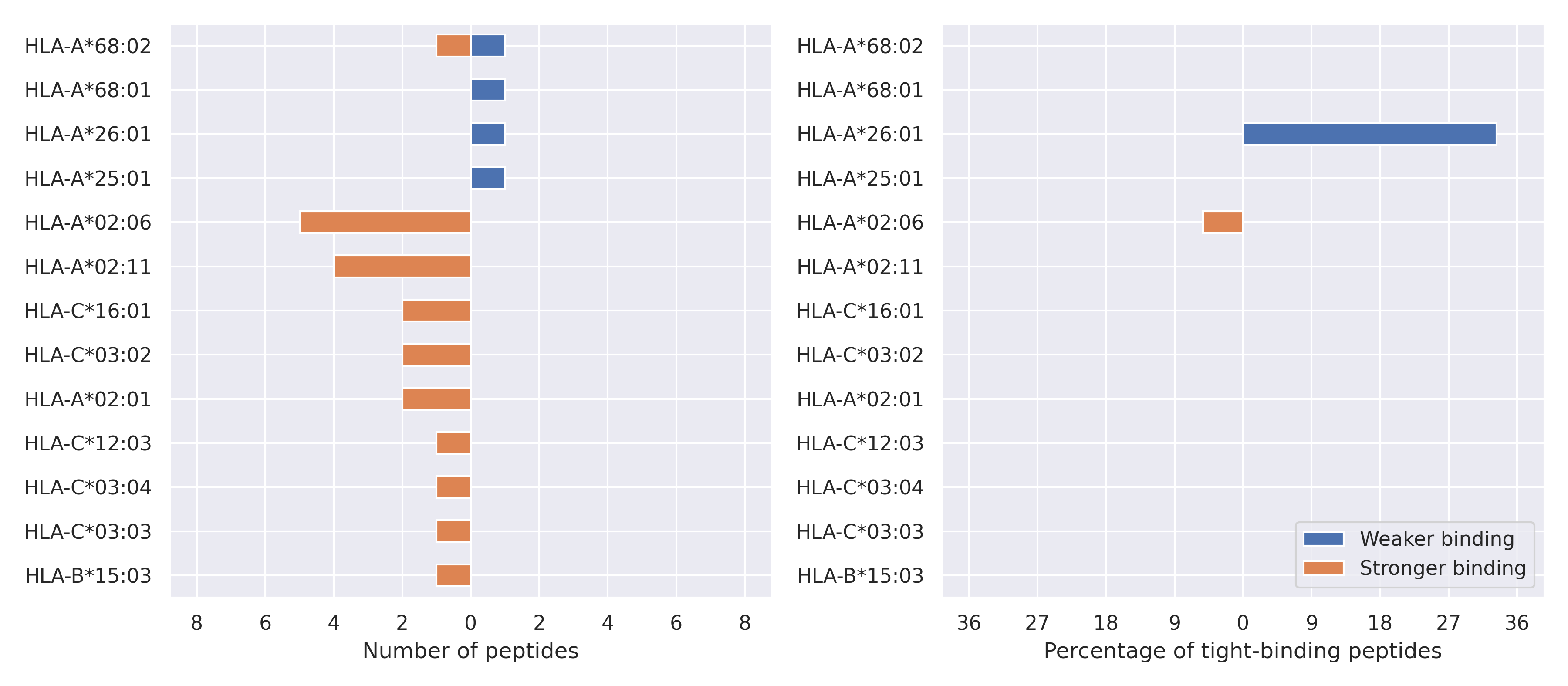
T223I
Reference KDCVVLHSYFTSDYYQLYSTQLSTDTGVEHVTFFIYNKIVDEPEEHVQIHT
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated KDCVVLHSYFTSDYYQLYSTQLSTDIGVEHVTFFIYNKIVDEPEEHVQIHT
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-C*03:03 | YSTQLSTDT | YSTQLSTDI | 12349 | 241 |
| HLA-C*03:04 | YSTQLSTDT | YSTQLSTDI | 12349 | 241 |
| HLA-C*12:03 | YSTQLSTDT | YSTQLSTDI | 11751 | 430 |
| HLA-C*16:01 | YSTQLSTDT | YSTQLSTDI | 7819 | 294 |
| HLA-C*03:02 | YSTQLSTDT | YSTQLSTDI | 6466 | 347 |
| HLA-A*02:06 | YQLYSTQLSTDT | YQLYSTQLSTDI | 4179 | 287 |
| HLA-A*02:11 | QLYSTQLSTDT | QLYSTQLSTDI | 5151 | 437 |
| HLA-A*68:01 | TGVEHVTFFIYNK | IGVEHVTFFIYNK | 213 | 1427 |
| HLA-A*68:02 | LSTDTGVEHV | LSTDIGVEHV | 1765 | 265 |
| HLA-A*68:02 | TGVEHVTFFI | IGVEHVTFFI | 212 | 1093 |
| HLA-A*02:01 | TQLSTDTGV | TQLSTDIGV | 1084 | 283 |
| HLA-A*02:01 | YSTQLSTDTGV | YSTQLSTDIGV | 1288 | 379 |
| HLA-A*02:11 | STQLSTDTGV | STQLSTDIGV | 1485 | 443 |
| HLA-A*25:01 | DTGVEHVTFF | DIGVEHVTFF | 180 | 596 |
| HLA-A*02:11 | TQLSTDTGV | TQLSTDIGV | 436 | 133 |
| HLA-A*02:06 | STQLSTDTGV | STQLSTDIGV | 264 | 83 |
| HLA-A*02:11 | YSTQLSTDTGV | YSTQLSTDIGV | 1184 | 381 |
| HLA-A*26:01 | DTGVEHVTFF | DIGVEHVTFF | 37 | 108 |
| HLA-A*02:06 | YSTQLSTDTGV | YSTQLSTDIGV | 157 | 56 |
| HLA-C*03:02 | TGVEHVTFF | IGVEHVTFF | 237 | 88 |
| HLA-A*02:06 | TQLSTDTGV | TQLSTDIGV | 70 | 26 |
| HLA-B*15:03 | TGVEHVTFF | IGVEHVTFF | 305 | 115 |
| HLA-A*02:06 | LYSTQLSTDTGV | LYSTQLSTDIGV | 855 | 330 |
| HLA-C*16:01 | TGVEHVTFF | IGVEHVTFF | 392 | 185 |
NS9b protein
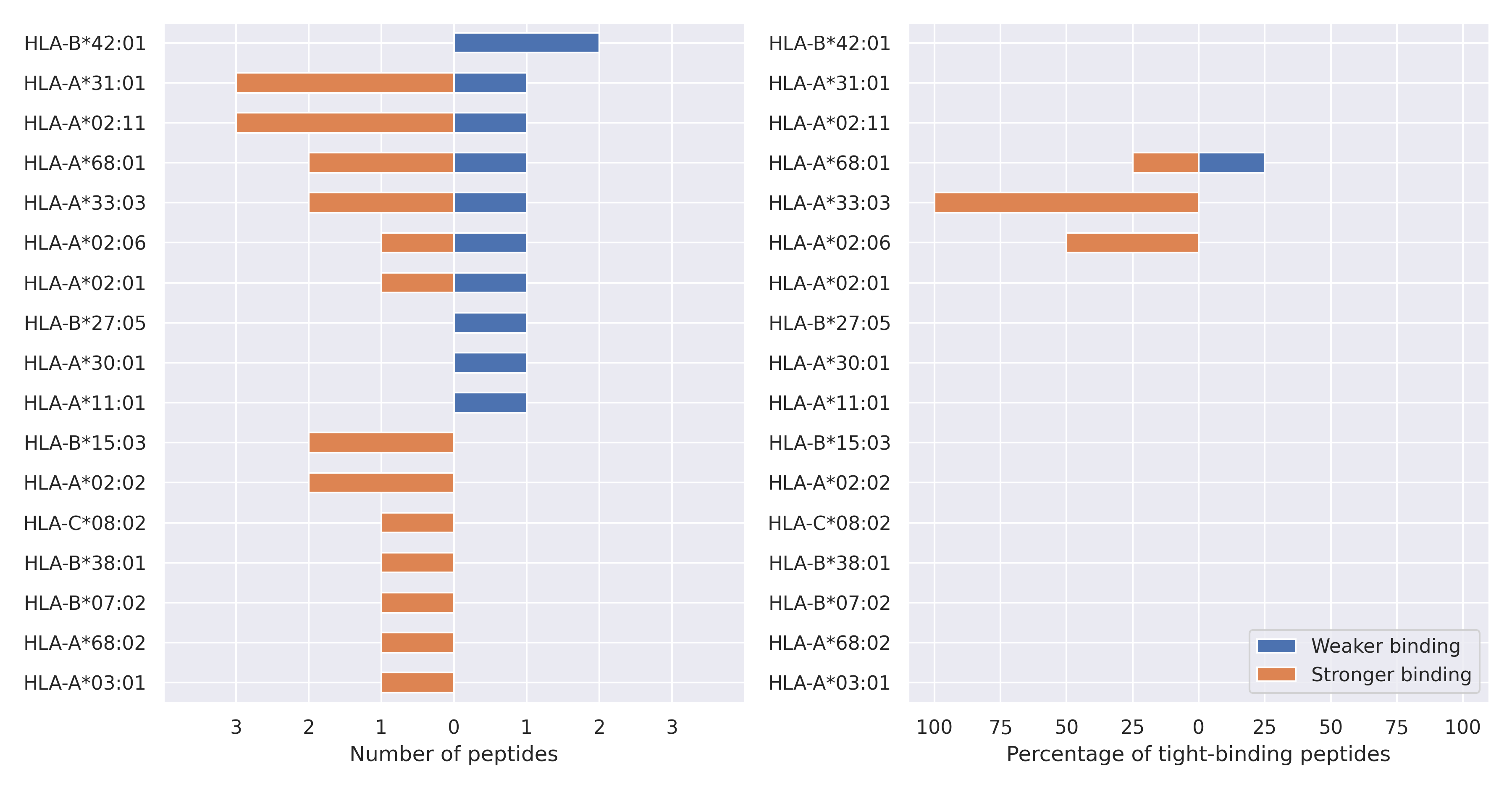
P10S
Reference MDPKISEMHPALRLVDPQIQLAVTRMENAVGRDQN
||||||||| |||||||||||||||||||||||||
Mutated MDPKISEMHSALRLVDPQIQLAVTRM‑‑‑VGRDQN
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-B*42:01 | HPALRLVDPQIQL | HSALRLVDPQIQL | 148 | 15057 |
| HLA-B*42:01 | HPALRLVDPQI | HSALRLVDPQI | 297 | 15937 |
| HLA-A*02:11 | PALRLVDPQIQL | SALRLVDPQIQL | 3683 | 325 |
| HLA-B*38:01 | MHPALRLVDPQI | MHSALRLVDPQI | 3064 | 395 |
| HLA-A*02:02 | PALRLVDPQIQL | SALRLVDPQIQL | 2220 | 388 |
| HLA-A*02:01 | KISEMHPAL | KISEMHSAL | 139 | 774 |
| HLA-A*02:06 | KISEMHPAL | KISEMHSAL | 62 | 310 |
| HLA-B*07:02 | KISEMHPAL | KISEMHSAL | 436 | 134 |
| HLA-A*02:11 | KISEMHPAL | KISEMHSAL | 14 | 42 |
| HLA-C*08:02 | ISEMHPAL | ISEMHSAL | 896 | 414 |
| HLA-B*15:03 | SEMHPALRL | SEMHSALRL | 493 | 245 |
ENA 27-29 deletion
Reference DPKISEMHPALRLVDPQIQLAVTRMENAVGRDQNNVGPKVYPIILRLGSPLSL
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated DPKISEMHSALRLVDPQIQLAVTRM‑‑‑VGRDQNNVGPKVYPIILRLGSPLSL
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-A*68:01 | VTRMENAVGR | VTRMVGR | 28 | - |
| HLA-A*31:01 | VTRMENAVGR | VTRMVGR | 113 | - |
| HLA-A*03:01 | RMENAVGRDQNNVGPK | RMVGRDQNNVGPK | - | 168 |
| HLA-A*33:03 | VTRMENAVGR | VTRMVGR | 218 | - |
| HLA-B*27:05 | TRMENAVGR | TRMVGR | 260 | - |
| HLA-A*11:01 | VTRMENAVGR | VTRMVGR | 268 | - |
| HLA-A*30:01 | VTRMENAVG | VTRMVG | 395 | - |
| HLA-A*68:02 | MENAVGRDQNNV | MVGRDQNNV | 13036 | 480 |
| HLA-A*33:03 | LAVTRMENAVGR | LAVTRMVGR | 927 | 45 |
| HLA-A*02:11 | IQLAVTRMENAV | IQLAVTRMV | 1418 | 70 |
| HLA-A*31:01 | LAVTRMENAVGR | LAVTRMVGR | 1898 | 115 |
| HLA-A*02:02 | RMENAVGRDQNNV | RMVGRDQNNV | 3078 | 195 |
| HLA-A*68:01 | QLAVTRMENAVGR | QLAVTRMVGR | 1673 | 108 |
| HLA-B*15:03 | IQLAVTRMENAV | IQLAVTRMV | 2775 | 183 |
| HLA-A*33:03 | QLAVTRMENAVGR | QLAVTRMVGR | 1778 | 188 |
| HLA-A*02:11 | RMENAVGRDQNNV | RMVGRDQNNV | 1928 | 213 |
| HLA-A*31:01 | IQLAVTRMENAVGR | IQLAVTRMVGR | 1758 | 236 |
| HLA-A*31:01 | QLAVTRMENAVGR | QLAVTRMVGR | 2442 | 329 |
| HLA-A*68:01 | LAVTRMENAVGR | LAVTRMVGR | 207 | 32 |
| HLA-A*02:06 | IQLAVTRMENAV | IQLAVTRMV | 263 | 47 |
| HLA-A*02:01 | IQLAVTRMENAV | IQLAVTRMV | 2387 | 463 |
NS9c protein

G50N
Reference AAVKPLLVPHHVVATVQEIQLQAAVGELLLLEWLAMAVMLLLLCCCLTD
||||||||||||||||||||||||| |||||||||||||||||||||||
Mutated AAVKPLLVPHHVVATVQEIQLQAAVNELLLLEWLAMAVMLLLLCCCLTD
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-B*18:01 | GELLLLEWLAM | NELLLLEWLAM | 4979 | 247 |
| HLA-B*18:01 | GELLLLEW | NELLLLEW | 3915 | 401 |
| HLA-B*40:01 | GELLLLEWL | NELLLLEWL | 28 | 140 |
| HLA-A*02:11 | QLQAAVGEL | QLQAAVNEL | 120 | 27 |
| HLA-A*02:11 | IQLQAAVGEL | IQLQAAVNEL | 338 | 81 |
| HLA-B*40:02 | GELLLLEWL | NELLLLEWL | 52 | 204 |
| HLA-B*40:02 | GELLLLEWLAM | NELLLLEWLAM | 201 | 772 |
| HLA-A*02:01 | QLQAAVGEL | QLQAAVNEL | 1290 | 358 |
| HLA-A*02:02 | QLQAAVGEL | QLQAAVNEL | 92 | 28 |
| HLA-B*58:01 | VGELLLLEW | VNELLLLEW | 330 | 1071 |
| HLA-A*02:02 | IQLQAAVGEL | IQLQAAVNEL | 170 | 58 |
| HLA-A*02:01 | LQAAVGELL | LQAAVNELL | 938 | 352 |
| HLA-A*02:02 | LQAAVGELL | LQAAVNELL | 44 | 17 |
| HLA-B*40:06 | GELLLLEWLA | NELLLLEWLA | 268 | 677 |
| HLA-B*40:02 | GELLLLEWLA | NELLLLEWLA | 475 | 1195 |
| HLA-A*02:11 | LQAAVGELL | LQAAVNELL | 386 | 154 |
| HLA-A*02:01 | IQLQAAVGEL | IQLQAAVNEL | 1138 | 475 |
| HLA-A*02:01 | QLQAAVGELL | QLQAAVNELL | 961 | 403 |
| HLA-A*02:01 | GELLLLEWLAMA | NELLLLEWLAMA | 109 | 258 |
| HLA-A*02:02 | QLQAAVGELL | QLQAAVNELL | 35 | 15 |
| HLA-A*02:02 | IQLQAAVGELL | IQLQAAVNELL | 724 | 313 |
| HLA-A*02:11 | QLQAAVGELL | QLQAAVNELL | 303 | 131 |
| HLA-B*58:01 | AVGELLLLEW | AVNELLLLEW | 196 | 87 |
| HLA-A*02:11 | GELLLLEWLAMA | NELLLLEWLAMA | 29 | 61 |
| HLA-B*57:01 | AVGELLLLEW | AVNELLLLEW | 432 | 206 |
| HLA-A*68:02 | EIQLQAAVGELL | EIQLQAAVNELL | 697 | 346 |
NSP1 protein
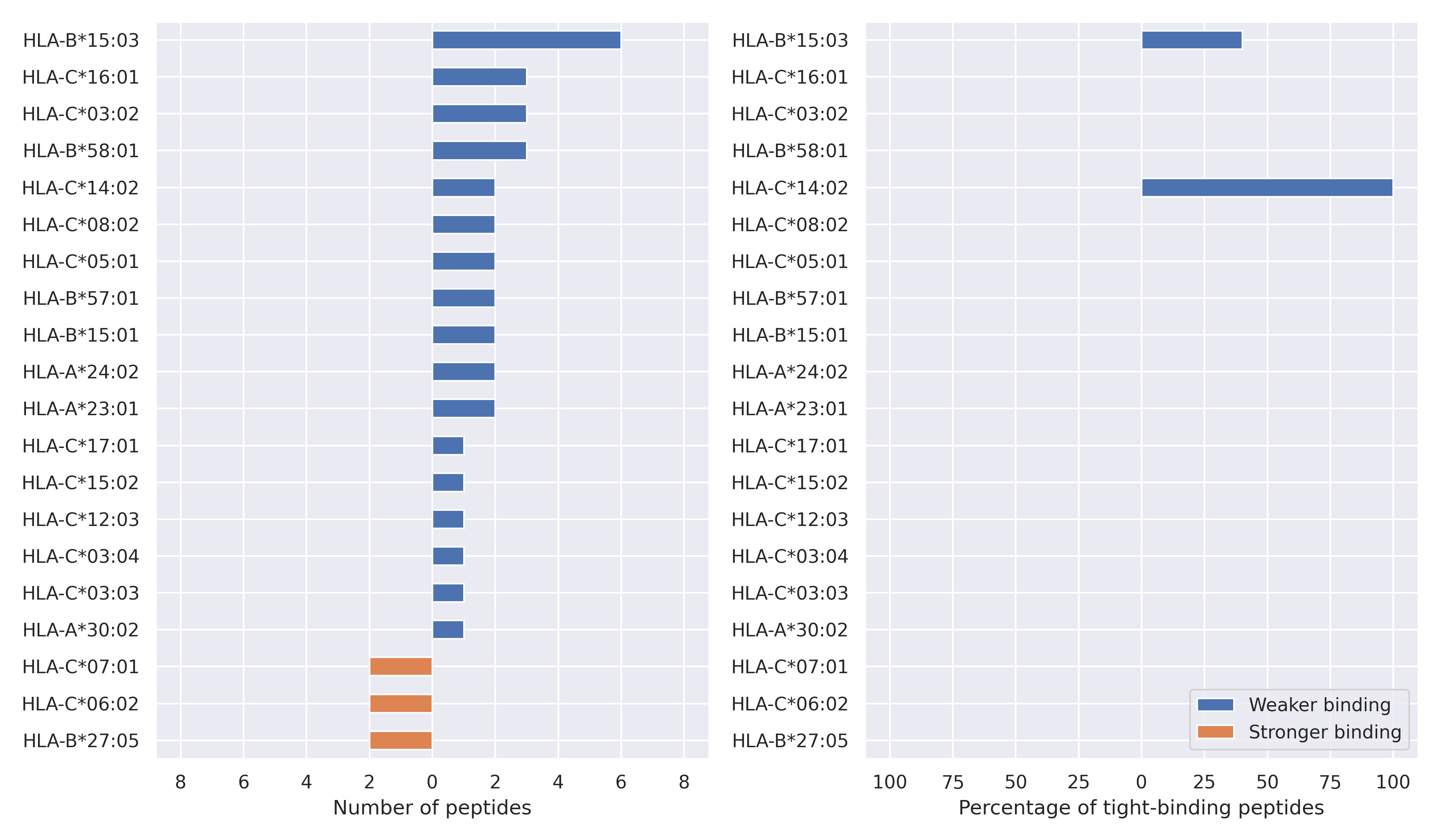
S135R
Reference HVGEIPVAYRKVLLRKNGNKGAGGHSYGADLKSFDLGDELGTDPYEDFQEN
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated HVGEIPVAYRKVLLRKNGNKGAGGHRYGADL‑‑‑DLGDELGTDPYEDFQEN
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-C*14:02 | SYGADLKSF | RYGADL | 37 | - |
| HLA-B*15:03 | HSYGADLKSF | HRYGADL | 41 | - |
| HLA-B*58:01 | HSYGADLKSF | HRYGADL | 59 | - |
| HLA-A*24:02 | SYGADLKSF | RYGADL | 84 | - |
| HLA-B*15:01 | HSYGADLKSF | HRYGADL | 94 | - |
| HLA-C*03:02 | HSYGADLKSF | HRYGADL | 94 | - |
| HLA-A*23:01 | SYGADLKSF | RYGADL | 129 | - |
| HLA-B*57:01 | HSYGADLKSF | HRYGADL | 158 | - |
| HLA-C*16:01 | HSYGADLKSF | HRYGADL | 329 | - |
| HLA-B*27:05 | HSYGADLKSFDL | HRYGADLDL | 23268 | 225 |
| HLA-C*06:02 | HSYGADLKSFDL | HRYGADLDL | 22732 | 410 |
| HLA-C*07:01 | HSYGADLKSFDL | HRYGADLDL | 18869 | 378 |
| HLA-B*15:03 | RKNGNKGAGGHSY | RKNGNKGAGGHRY | 183 | 1186 |
| HLA-B*15:03 | NKGAGGHSY | NKGAGGHRY | 129 | 774 |
| HLA-B*15:03 | GNKGAGGHSY | GNKGAGGHRY | 478 | 2611 |
KSF 141-143 deletion
Reference VAYRKVLLRKNGNKGAGGHSYGADLKSFDLGDELGTDPYEDFQENWNTKHSSG
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated VAYRKVLLRKNGNKGAGGHRYGADL‑‑‑DLGDELGTDPYEDFQENWNTKHSSG
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-C*14:02 | SYGADLKSF | RYGADL | 37 | - |
| HLA-B*15:03 | HSYGADLKSF | HRYGADL | 41 | - |
| HLA-B*58:01 | HSYGADLKSF | HRYGADL | 59 | - |
| HLA-A*24:02 | SYGADLKSF | RYGADL | 84 | - |
| HLA-B*15:01 | HSYGADLKSF | HRYGADL | 94 | - |
| HLA-C*03:02 | HSYGADLKSF | HRYGADL | 94 | - |
| HLA-A*23:01 | SYGADLKSF | RYGADL | 129 | - |
| HLA-C*03:03 | KSFDLGDEL | DLGDEL | 148 | - |
| HLA-C*03:04 | KSFDLGDEL | DLGDEL | 148 | - |
| HLA-B*57:01 | HSYGADLKSF | HRYGADL | 158 | - |
| HLA-C*16:01 | KSFDLGDEL | DLGDEL | 170 | - |
| HLA-A*30:02 | KSFDLGDELGTDPY | - | 237 | - |
| HLA-C*17:01 | KSFDLGDEL | DLGDEL | 271 | - |
| HLA-C*05:01 | YGADLKSFDL | YGADLDL | 275 | - |
| HLA-C*03:02 | KSFDLGDEL | DLGDEL | 283 | - |
| HLA-C*08:02 | YGADLKSFDL | YGADLDL | 297 | - |
| HLA-B*15:03 | KSFDLGDEL | DLGDEL | 309 | - |
| HLA-C*16:01 | HSYGADLKSF | HRYGADL | 329 | - |
| HLA-C*05:01 | GADLKSFDL | GADLDL | 343 | - |
| HLA-C*12:03 | KSFDLGDEL | DLGDEL | 358 | - |
| HLA-C*08:02 | GADLKSFDL | GADLDL | 361 | - |
| HLA-B*58:01 | KSFDLGDEL | DLGDEL | 366 | - |
| HLA-C*15:02 | KSFDLGDEL | DLGDEL | 414 | - |
| HLA-B*27:05 | HSYGADLKSFDL | HRYGADLDL | 23268 | 225 |
| HLA-C*06:02 | HSYGADLKSFDL | HRYGADLDL | 22732 | 410 |
| HLA-C*07:01 | HSYGADLKSFDL | HRYGADLDL | 18869 | 378 |
NSP3 protein
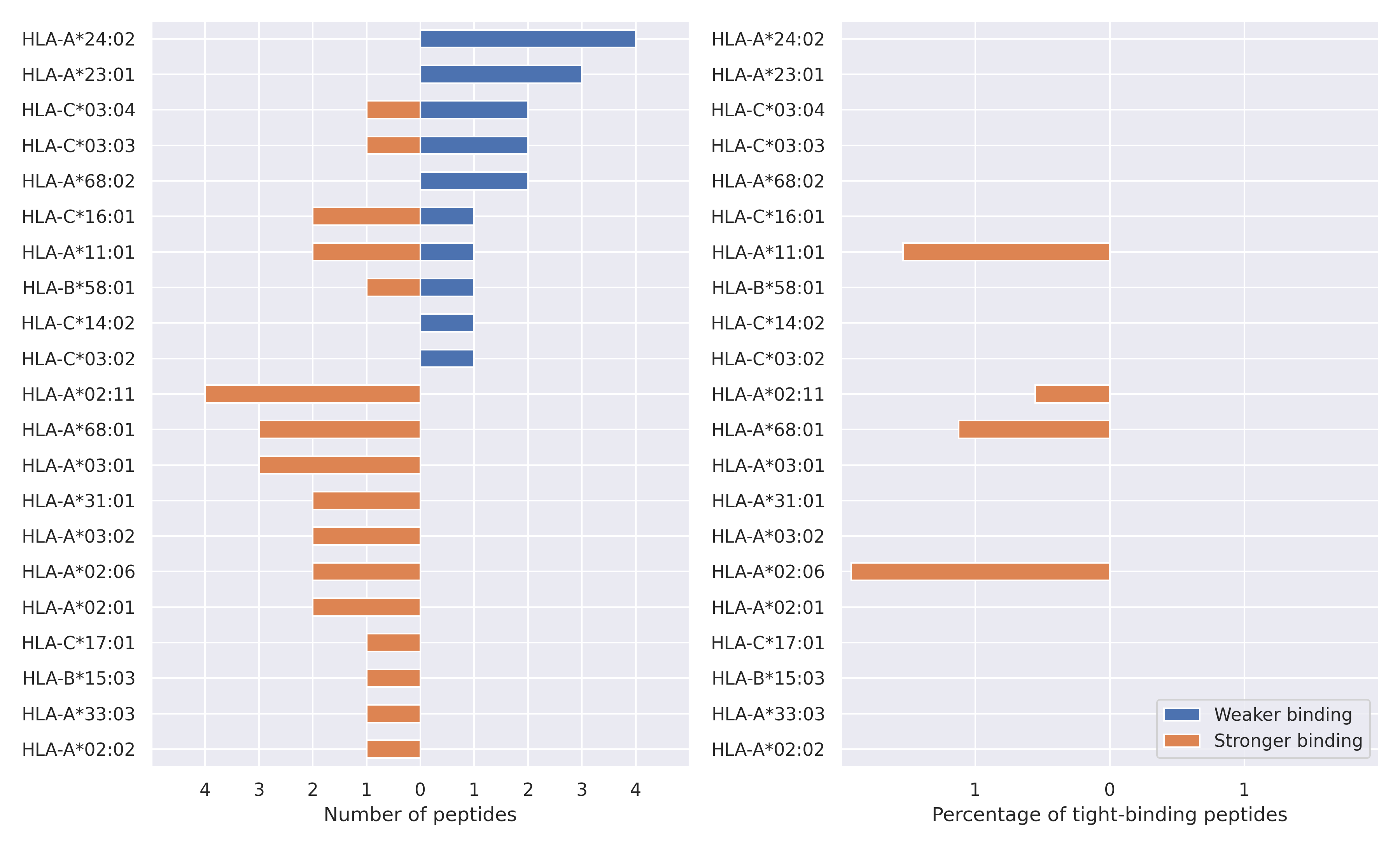
T24I
Reference APTKVTFGDDTVIEVQGYKSVNITFELDERIDKVLNEKCSAYTVELGTE
||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated APTKVTFGDDTVIEVQGYKSVNIIFELDERIDKVLNEKCSAYTVELGTE
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-B*58:01 | ITFELDERI | IIFELDERI | 270 | 4454 |
| HLA-B*15:03 | VQGYKSVNIT | VQGYKSVNII | 3702 | 395 |
| HLA-A*02:01 | ITFELDERI | IIFELDERI | 1637 | 223 |
| HLA-A*02:11 | ITFELDERI | IIFELDERI | 261 | 37 |
| HLA-A*02:02 | ITFELDERI | IIFELDERI | 428 | 65 |
| HLA-A*24:02 | VQGYKSVNITF | VQGYKSVNIIF | 194 | 811 |
| HLA-A*24:02 | IEVQGYKSVNITF | IEVQGYKSVNIIF | 366 | 1499 |
| HLA-A*68:02 | ITFELDERI | IIFELDERI | 253 | 1036 |
| HLA-A*68:01 | NITFELDER | NIIFELDER | 68 | 20 |
| HLA-A*33:03 | NITFELDER | NIIFELDER | 874 | 262 |
| HLA-A*23:01 | IEVQGYKSVNITF | IEVQGYKSVNIIF | 191 | 635 |
| HLA-C*03:03 | YKSVNITFEL | YKSVNIIFEL | 305 | 1007 |
| HLA-C*03:04 | YKSVNITFEL | YKSVNIIFEL | 305 | 1007 |
| HLA-A*23:01 | VIEVQGYKSVNITF | VIEVQGYKSVNIIF | 413 | 1337 |
| HLA-A*23:01 | VQGYKSVNITF | VQGYKSVNIIF | 111 | 353 |
| HLA-A*02:11 | KSVNITFEL | KSVNIIFEL | 169 | 55 |
| HLA-C*14:02 | GYKSVNITF | GYKSVNIIF | 53 | 160 |
| HLA-A*11:01 | ITFELDERIDK | IIFELDERIDK | 446 | 1259 |
| HLA-A*02:01 | KSVNITFEL | KSVNIIFEL | 863 | 307 |
| HLA-C*03:02 | YKSVNITFEL | YKSVNIIFEL | 299 | 784 |
| HLA-A*68:01 | VNITFELDER | VNIIFELDER | 1032 | 394 |
| HLA-A*02:06 | KSVNITFEL | KSVNIIFEL | 80 | 31 |
| HLA-C*16:01 | YKSVNITFEL | YKSVNIIFEL | 310 | 793 |
| HLA-A*68:02 | NITFELDERI | NIIFELDERI | 209 | 533 |
| HLA-A*24:02 | GYKSVNITF | GYKSVNIIF | 83 | 192 |
| HLA-A*02:11 | ITFELDERIDKV | IIFELDERIDKV | 397 | 175 |
| HLA-A*68:01 | SVNITFELDER | SVNIIFELDER | 405 | 180 |
| HLA-A*24:02 | QGYKSVNITF | QGYKSVNIIF | 208 | 455 |
| HLA-A*02:06 | ITFELDERI | IIFELDERI | 402 | 185 |
| HLA-A*02:11 | YKSVNITFEL | YKSVNIIFEL | 715 | 337 |
| HLA-C*03:03 | KSVNITFEL | KSVNIIFEL | 115 | 233 |
| HLA-C*03:04 | KSVNITFEL | KSVNIIFEL | 115 | 233 |
G489S
Reference DAPYIVGDVVQEGVLTAVVIPTKKAGGTTEMLAKALRKVPTDNYITTYPGQ
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated DAPYIVGDVVQEGVLTAVVIPTKKASGTTEMLAKALRKVPTDNYITTYPGQ
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-A*11:01 | AGGTTEMLAK | ASGTTEMLAK | 1254 | 37 |
| HLA-A*03:01 | AGGTTEMLAK | ASGTTEMLAK | 3665 | 277 |
| HLA-A*03:02 | AGGTTEMLAK | ASGTTEMLAK | 2689 | 228 |
| HLA-C*03:03 | KAGGTTEML | KASGTTEML | 2418 | 212 |
| HLA-C*03:04 | KAGGTTEML | KASGTTEML | 2418 | 212 |
| HLA-C*16:01 | KAGGTTEML | KASGTTEML | 1012 | 116 |
| HLA-C*17:01 | KAGGTTEML | KASGTTEML | 1802 | 287 |
| HLA-A*31:01 | AGGTTEMLAKALR | ASGTTEMLAKALR | 787 | 133 |
| HLA-B*58:01 | KAGGTTEML | KASGTTEML | 2117 | 440 |
| HLA-C*16:01 | KAGGTTEM | KASGTTEM | 863 | 191 |
| HLA-A*03:02 | GGTTEMLAKALRK | SGTTEMLAKALRK | 1693 | 453 |
| HLA-A*11:01 | KAGGTTEMLAK | KASGTTEMLAK | 589 | 203 |
| HLA-A*03:01 | KAGGTTEMLAK | KASGTTEMLAK | 1236 | 449 |
| HLA-A*03:01 | GGTTEMLAKALRK | SGTTEMLAKALRK | 467 | 181 |
| HLA-A*31:01 | GGTTEMLAKALR | SGTTEMLAKALR | 677 | 335 |
NSP4 protein
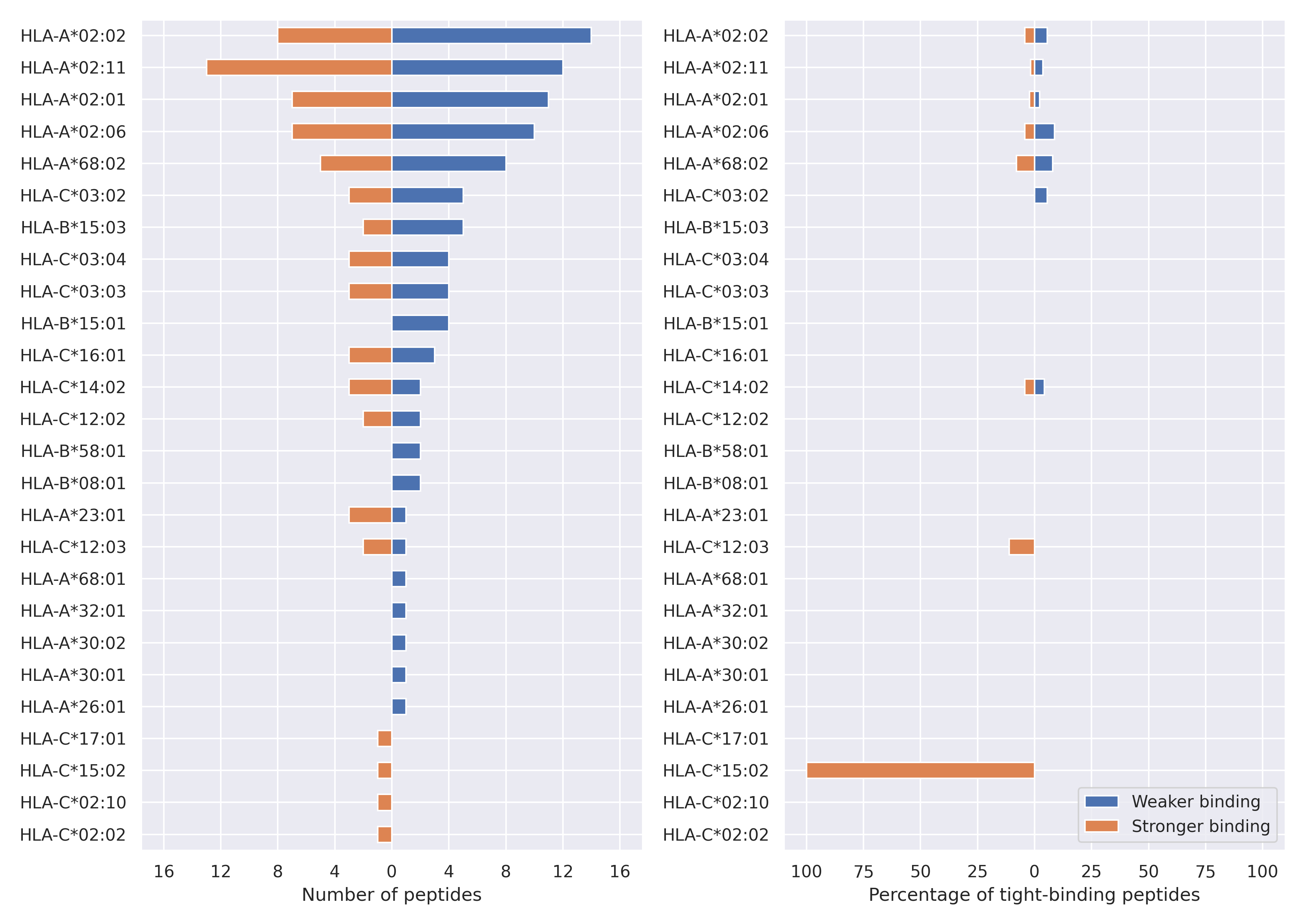
L264F
Reference GRWVLNNDYYRSLPGVFCGVDAVNLLTNMFTPLIQPIGALDISASIVAGGI
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated GRWVLNNDYYRSLPGVFCGVDAVNLFTNMFTPLIQPIGALDISASIVAGGI
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-A*02:11 | LLTNMFTPL | LFTNMFTPL | 5 | 2700 |
| HLA-A*02:01 | LLTNMFTPL | LFTNMFTPL | 16 | 6477 |
| HLA-A*02:02 | LLTNMFTPL | LFTNMFTPL | 5 | 1518 |
| HLA-A*02:06 | LLTNMFTPL | LFTNMFTPL | 28 | 3590 |
| HLA-A*02:11 | LLTNMFTPLI | LFTNMFTPLI | 17 | 1231 |
| HLA-A*02:01 | LLTNMFTPLI | LFTNMFTPLI | 55 | 3749 |
| HLA-C*14:02 | LLTNMFTPL | LFTNMFTPL | 453 | 7 |
| HLA-A*02:02 | LLTNMFTPLI | LFTNMFTPLI | 22 | 967 |
| HLA-B*15:01 | LLTNMFTPL | LFTNMFTPL | 228 | 5949 |
| HLA-C*03:03 | LTNMFTPL | FTNMFTPL | 1113 | 61 |
| HLA-C*03:04 | LTNMFTPL | FTNMFTPL | 1113 | 61 |
| HLA-A*23:01 | VFCGVDAVNLL | VFCGVDAVNLF | 2671 | 151 |
| HLA-A*02:02 | LLTNMFTPLIQPI | LFTNMFTPLIQPI | 294 | 4718 |
| HLA-A*02:02 | GVFCGVDAVNLL | GVFCGVDAVNLF | 439 | 6440 |
| HLA-C*14:02 | NLLTNMFTPL | NLFTNMFTPL | 4861 | 337 |
| HLA-B*15:03 | CGVDAVNLL | CGVDAVNLF | 6087 | 490 |
| HLA-A*02:11 | LLTNMFTPLIQPI | LFTNMFTPLIQPI | 297 | 3332 |
| HLA-C*03:02 | LTNMFTPL | FTNMFTPL | 829 | 77 |
| HLA-C*03:03 | LTNMFTPLI | FTNMFTPLI | 2207 | 207 |
| HLA-C*03:04 | LTNMFTPLI | FTNMFTPLI | 2207 | 207 |
| HLA-A*02:01 | LTNMFTPLI | FTNMFTPLI | 1097 | 103 |
| HLA-A*02:11 | LTNMFTPLI | FTNMFTPLI | 136 | 13 |
| HLA-C*12:03 | LTNMFTPLI | FTNMFTPLI | 430 | 42 |
| HLA-A*02:06 | LLTNMFTPLI | LFTNMFTPLI | 136 | 1345 |
| HLA-A*02:06 | LTNMFTPLI | FTNMFTPLI | 223 | 23 |
| HLA-C*12:02 | LTNMFTPLI | FTNMFTPLI | 1404 | 157 |
| HLA-A*02:02 | LTNMFTPLI | FTNMFTPLI | 364 | 41 |
| HLA-A*68:02 | LTNMFTPLIQPI | FTNMFTPLIQPI | 1086 | 129 |
| HLA-C*12:03 | LTNMFTPL | FTNMFTPL | 2677 | 324 |
| HLA-C*03:02 | LTNMFTPLI | FTNMFTPLI | 716 | 87 |
| HLA-C*02:02 | LTNMFTPLI | FTNMFTPLI | 1268 | 158 |
| HLA-C*02:10 | LTNMFTPLI | FTNMFTPLI | 1268 | 158 |
| HLA-C*17:01 | LTNMFTPLI | FTNMFTPLI | 599 | 86 |
| HLA-A*68:02 | LTNMFTPLI | FTNMFTPLI | 82 | 12 |
| HLA-C*03:03 | LLTNMFTPL | LFTNMFTPL | 253 | 1728 |
| HLA-C*03:04 | LLTNMFTPL | LFTNMFTPL | 253 | 1728 |
| HLA-C*12:02 | LTNMFTPL | FTNMFTPL | 3002 | 450 |
| HLA-B*15:03 | LLTNMFTPL | LFTNMFTPL | 108 | 704 |
| HLA-A*68:02 | NLLTNMFTPL | NLFTNMFTPL | 2741 | 480 |
| HLA-C*14:02 | LTNMFTPL | FTNMFTPL | 2589 | 467 |
| HLA-A*02:06 | LTNMFTPLIQPI | FTNMFTPLIQPI | 2171 | 394 |
| HLA-C*16:01 | LTNMFTPL | FTNMFTPL | 293 | 61 |
| HLA-C*16:01 | LTNMFTPLI | FTNMFTPLI | 267 | 56 |
| HLA-A*30:01 | LTNMFTPLI | FTNMFTPLI | 144 | 648 |
| HLA-A*02:02 | AVNLLTNMFTPL | AVNLFTNMFTPL | 249 | 948 |
| HLA-B*08:01 | LLTNMFTPL | LFTNMFTPL | 271 | 983 |
| HLA-A*02:11 | LTNMFTPLIQPI | FTNMFTPLIQPI | 1413 | 409 |
| HLA-C*03:02 | LLTNMFTPL | LFTNMFTPL | 307 | 951 |
| HLA-A*02:02 | NLLTNMFTPLI | NLFTNMFTPLI | 658 | 224 |
| HLA-A*02:02 | NLLTNMFTPL | NLFTNMFTPL | 58 | 21 |
| HLA-C*15:02 | LTNMFTPLI | FTNMFTPLI | 67 | 25 |
| HLA-A*02:11 | AVNLLTNMFTPL | AVNLFTNMFTPL | 390 | 1009 |
| HLA-A*02:02 | VNLLTNMFTPL | VNLFTNMFTPL | 375 | 919 |
| HLA-A*02:11 | NLLTNMFTP | NLFTNMFTP | 1166 | 476 |
| HLA-B*15:03 | AVNLLTNMF | AVNLFTNMF | 390 | 172 |
| HLA-A*02:11 | NLLTNMFTPLIQPI | NLFTNMFTPLIQPI | 274 | 129 |
T327I
Reference MRFRRAFGEYSHVVAFNTLLFLMSFTVLCLTPVYSFLPGVYSVIYLYLTFY
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated MRFRRAFGEYSHVVAFNTLLFLMSFIVLCLTPVYSFLPGVYSVIYLYLTFY
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-A*02:11 | NTLLFLMSFT | NTLLFLMSFI | 1059 | 51 |
| HLA-A*68:02 | NTLLFLMSFT | NTLLFLMSFI | 741 | 41 |
| HLA-A*02:11 | FNTLLFLMSFT | FNTLLFLMSFI | 1509 | 110 |
| HLA-A*02:11 | TLLFLMSFT | TLLFLMSFI | 82 | 6 |
| HLA-A*68:02 | TLLFLMSFT | TLLFLMSFI | 3858 | 286 |
| HLA-A*68:02 | FTVLCLTPVYSFL | FIVLCLTPVYSFL | 129 | 1580 |
| HLA-A*02:11 | AFNTLLFLMSFT | AFNTLLFLMSFI | 4598 | 420 |
| HLA-A*68:02 | FTVLCLTPV | FIVLCLTPV | 8 | 83 |
| HLA-A*68:02 | SFTVLCLTPV | SFIVLCLTPV | 63 | 597 |
| HLA-A*02:01 | TLLFLMSFT | TLLFLMSFI | 239 | 26 |
| HLA-A*02:01 | NTLLFLMSFT | NTLLFLMSFI | 1593 | 179 |
| HLA-A*02:02 | TLLFLMSFT | TLLFLMSFI | 235 | 28 |
| HLA-B*15:01 | LLFLMSFTVL | LLFLMSFIVL | 229 | 1839 |
| HLA-A*02:11 | LFLMSFTV | LFLMSFIV | 279 | 2222 |
| HLA-A*02:02 | NTLLFLMSFT | NTLLFLMSFI | 1462 | 186 |
| HLA-A*02:11 | LLFLMSFT | LLFLMSFI | 2935 | 377 |
| HLA-A*32:01 | LLFLMSFTV | LLFLMSFIV | 369 | 2631 |
| HLA-A*02:06 | LLFLMSFTV | LLFLMSFIV | 34 | 231 |
| HLA-C*12:03 | FTVLCLTPV | FIVLCLTPV | 161 | 1041 |
| HLA-C*14:02 | LFLMSFTVL | LFLMSFIVL | 18 | 112 |
| HLA-A*02:11 | AFNTLLFLMSFTV | AFNTLLFLMSFIV | 74 | 448 |
| HLA-C*16:01 | FTVLCLTPVY | FIVLCLTPVY | 395 | 2359 |
| HLA-A*02:06 | TLLFLMSFT | TLLFLMSFI | 397 | 68 |
| HLA-A*02:01 | LLFLMSFTV | LLFLMSFIV | 9 | 50 |
| HLA-A*02:11 | VAFNTLLFLMSFTV | VAFNTLLFLMSFIV | 225 | 1206 |
| HLA-A*02:01 | AFNTLLFLMSFTV | AFNTLLFLMSFIV | 357 | 1842 |
| HLA-A*68:02 | NTLLFLMSFTV | NTLLFLMSFIV | 106 | 539 |
| HLA-A*02:11 | NTLLFLMSFTV | NTLLFLMSFIV | 59 | 300 |
| HLA-A*02:01 | NTLLFLMSFTV | NTLLFLMSFIV | 212 | 1074 |
| HLA-C*03:03 | FTVLCLTPV | FIVLCLTPV | 302 | 1526 |
| HLA-C*03:04 | FTVLCLTPV | FIVLCLTPV | 302 | 1526 |
| HLA-B*15:03 | LLFLMSFTVL | LLFLMSFIVL | 318 | 1563 |
| HLA-A*02:02 | LLFLMSFTV | LLFLMSFIV | 11 | 53 |
| HLA-A*02:06 | NTLLFLMSFT | NTLLFLMSFI | 945 | 197 |
| HLA-A*02:11 | SFTVLCLTPV | SFIVLCLTPV | 250 | 53 |
| HLA-A*02:11 | FNTLLFLMSFTV | FNTLLFLMSFIV | 23 | 108 |
| HLA-A*02:01 | FNTLLFLMSFTV | FNTLLFLMSFIV | 131 | 602 |
| HLA-C*03:02 | FTVLCLTPV | FIVLCLTPV | 198 | 902 |
| HLA-C*14:02 | FLMSFTVL | FLMSFIVL | 475 | 2138 |
| HLA-C*12:02 | FTVLCLTPV | FIVLCLTPV | 218 | 961 |
| HLA-C*03:03 | FLMSFTVL | FLMSFIVL | 309 | 1359 |
| HLA-C*03:04 | FLMSFTVL | FLMSFIVL | 309 | 1359 |
| HLA-A*26:01 | FTVLCLTPVY | FIVLCLTPVY | 176 | 760 |
| HLA-A*02:01 | SFTVLCLTPV | SFIVLCLTPV | 470 | 111 |
| HLA-B*15:01 | FLMSFTVLCL | FLMSFIVLCL | 455 | 1914 |
| HLA-B*15:03 | LFLMSFTVL | LFLMSFIVL | 353 | 1462 |
| HLA-A*68:01 | FTVLCLTPVY | FIVLCLTPVY | 485 | 1972 |
| HLA-A*02:02 | FNTLLFLMSFTV | FNTLLFLMSFIV | 186 | 704 |
| HLA-B*15:01 | LMSFTVLCL | LMSFIVLCL | 110 | 408 |
| HLA-C*03:02 | FLMSFTVL | FLMSFIVL | 352 | 1285 |
| HLA-A*02:02 | NTLLFLMSFTV | NTLLFLMSFIV | 424 | 1515 |
| HLA-A*02:11 | LLFLMSFTV | LLFLMSFIV | 2 | 7 |
| HLA-C*03:02 | FTVLCLTPVY | FIVLCLTPVY | 50 | 175 |
| HLA-B*15:03 | LMSFTVLCL | LMSFIVLCL | 54 | 186 |
| HLA-C*12:02 | FTVLCLTPVY | FIVLCLTPVY | 331 | 1137 |
| HLA-A*02:02 | LLFLMSFTVL | LLFLMSFIVL | 42 | 144 |
| HLA-A*68:02 | MSFTVLCLTPV | MSFIVLCLTPV | 34 | 114 |
| HLA-C*03:02 | LMSFTVLCL | LMSFIVLCL | 467 | 1520 |
| HLA-A*02:01 | LLFLMSFTVL | LLFLMSFIVL | 74 | 240 |
| HLA-A*02:11 | LLFLMSFTVL | LLFLMSFIVL | 25 | 80 |
| HLA-A*02:02 | SFTVLCLTPV | SFIVLCLTPV | 261 | 84 |
| HLA-A*02:02 | LFLMSFTVLCL | LFLMSFIVLCL | 73 | 216 |
| HLA-A*02:06 | LLFLMSFTVL | LLFLMSFIVL | 331 | 973 |
| HLA-C*16:01 | MSFTVLCL | MSFIVLCL | 171 | 496 |
| HLA-A*68:02 | FLMSFTVLCLTPV | FLMSFIVLCLTPV | 331 | 927 |
| HLA-A*68:02 | MSFTVLCLT | MSFIVLCLT | 68 | 186 |
| HLA-A*02:06 | MSFTVLCLT | MSFIVLCLT | 239 | 650 |
| HLA-A*02:11 | TVLCLTPVYSFL | IVLCLTPVYSFL | 533 | 199 |
| HLA-B*15:03 | FLMSFTVL | FLMSFIVL | 371 | 993 |
| HLA-A*02:02 | FLMSFTVLCL | FLMSFIVLCL | 3 | 8 |
| HLA-A*02:02 | LLFLMSFTVLCL | LLFLMSFIVLCL | 134 | 356 |
| HLA-A*02:01 | LLFLMSFTVLCL | LLFLMSFIVLCL | 237 | 602 |
| HLA-A*02:01 | LFLMSFTVLCL | LFLMSFIVLCL | 73 | 185 |
| HLA-A*02:06 | FLMSFTVLCL | FLMSFIVLCL | 15 | 38 |
| HLA-C*03:02 | TVLCLTPVY | IVLCLTPVY | 136 | 54 |
| HLA-A*23:01 | LFLMSFTVL | LFLMSFIVL | 411 | 1026 |
| HLA-A*02:06 | TLLFLMSFTV | TLLFLMSFIV | 47 | 116 |
| HLA-B*08:01 | FLMSFTVL | FLMSFIVL | 118 | 291 |
| HLA-A*02:06 | NTLLFLMSFTV | NTLLFLMSFIV | 373 | 906 |
| HLA-A*02:01 | TLLFLMSFTV | TLLFLMSFIV | 14 | 33 |
| HLA-A*02:06 | VAFNTLLFLMSFTV | VAFNTLLFLMSFIV | 479 | 1101 |
| HLA-A*02:06 | MSFTVLCLTPV | MSFIVLCLTPV | 48 | 110 |
| HLA-B*58:01 | MSFTVLCLT | MSFIVLCLT | 412 | 943 |
| HLA-A*02:01 | FLMSFTVLCL | FLMSFIVLCL | 4 | 9 |
| HLA-A*02:02 | FLMSFTVLCLTP | FLMSFIVLCLTP | 194 | 430 |
| HLA-A*02:11 | TLLFLMSFTV | TLLFLMSFIV | 5 | 11 |
| HLA-A*02:02 | FTVLCLTPV | FIVLCLTPV | 23 | 11 |
| HLA-A*02:01 | FTVLCLTPV | FIVLCLTPV | 25 | 12 |
| HLA-A*30:02 | FTVLCLTPVY | FIVLCLTPVY | 327 | 671 |
| HLA-A*02:02 | TVLCLTPVYSFL | IVLCLTPVYSFL | 176 | 86 |
| HLA-B*58:01 | MSFTVLCLTP | MSFIVLCLTP | 497 | 1015 |
| HLA-A*02:11 | FTVLCLTPV | FIVLCLTPV | 12 | 6 |
T492I
Reference ACCHLAKALNDFSNSGSDVLYQPPQTSITSAVLQ
||||||||||||||||||||||||| ||||||||
Mutated ACCHLAKALNDFSNSGSDVLYQPPQISITSAVLQ
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-C*16:01 | QTSITSAVL | QISITSAVL | 201 | 1829 |
| HLA-A*68:02 | QTSITSAVL | QISITSAVL | 206 | 1702 |
| HLA-A*02:06 | QPPQTSITSAV | QPPQISITSAV | 3097 | 393 |
| HLA-A*02:06 | LYQPPQTSITSAV | LYQPPQISITSAV | 1485 | 214 |
| HLA-A*02:06 | YQPPQTSITSAV | YQPPQISITSAV | 156 | 26 |
| HLA-A*02:01 | YQPPQTSITSAV | YQPPQISITSAV | 1613 | 341 |
| HLA-A*02:11 | YQPPQTSITSAV | YQPPQISITSAV | 498 | 120 |
| HLA-C*03:03 | TSITSAVL | ISITSAVL | 532 | 198 |
| HLA-C*03:04 | TSITSAVL | ISITSAVL | 532 | 198 |
| HLA-C*03:03 | QTSITSAVL | QISITSAVL | 311 | 831 |
| HLA-C*03:04 | QTSITSAVL | QISITSAVL | 311 | 831 |
| HLA-C*16:01 | TSITSAVL | ISITSAVL | 381 | 156 |
| HLA-A*02:01 | VLYQPPQTSI | VLYQPPQISI | 1084 | 474 |
| HLA-A*23:01 | VLYQPPQTSI | VLYQPPQISI | 979 | 462 |
| HLA-A*23:01 | LYQPPQTSI | LYQPPQISI | 278 | 132 |
NSP5 protein

P132H
Reference QPGQTFSVLACYNGSPSGVYQCAMRPNFTIKGSFLNGSCGSVGFNIDYDCV
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated QPGQTFSVLACYNGSPSGVYQCAMRHNFTIKGSFLNGSCGSVGFNIDYDCV
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-B*07:02 | RPNFTIKGSF | RHNFTIKGSF | 14 | 15756 |
| HLA-B*07:02 | RPNFTIKGSFL | RHNFTIKGSFL | 24 | 18650 |
| HLA-B*42:01 | RPNFTIKGSF | RHNFTIKGSF | 31 | 15129 |
| HLA-B*42:01 | RPNFTIKGSFL | RHNFTIKGSFL | 39 | 17143 |
| HLA-B*15:03 | PNFTIKGSF | HNFTIKGSF | 7728 | 117 |
| HLA-A*03:01 | GVYQCAMRP | GVYQCAMRH | 20799 | 417 |
| HLA-B*08:01 | CAMRPNFTI | CAMRHNFTI | 3487 | 242 |
| HLA-B*27:05 | MRPNFTIKG | MRHNFTIKG | 4762 | 366 |
| HLA-B*27:05 | MRPNFTIKGSFL | MRHNFTIKGSFL | 827 | 86 |
| HLA-B*15:03 | RPNFTIKGSF | RHNFTIKGSF | 432 | 54 |
| HLA-B*27:05 | MRPNFTIK | MRHNFTIK | 2132 | 276 |
| HLA-B*15:03 | MRPNFTIKGSF | MRHNFTIKGSF | 397 | 96 |
| HLA-C*07:02 | MRPNFTIKGSF | MRHNFTIKGSF | 361 | 1054 |
| HLA-B*27:05 | MRPNFTIKGSF | MRHNFTIKGSF | 218 | 78 |
| HLA-A*30:01 | QCAMRPNFTIK | QCAMRHNFTIK | 199 | 513 |
| HLA-A*68:02 | CAMRPNFTI | CAMRHNFTI | 199 | 493 |
| HLA-C*16:01 | CAMRPNFTI | CAMRHNFTI | 125 | 52 |
| HLA-A*30:01 | AMRPNFTIKG | AMRHNFTIKG | 123 | 279 |
| HLA-C*12:03 | CAMRPNFTI | CAMRHNFTI | 317 | 143 |
| HLA-A*30:01 | VYQCAMRPNFTIK | VYQCAMRHNFTIK | 316 | 683 |
| HLA-A*11:01 | AMRPNFTIK | AMRHNFTIK | 175 | 376 |
| HLA-A*11:01 | CAMRPNFTIK | CAMRHNFTIK | 42 | 88 |
| HLA-A*68:01 | CAMRPNFTIK | CAMRHNFTIK | 127 | 254 |
NSP6 protein
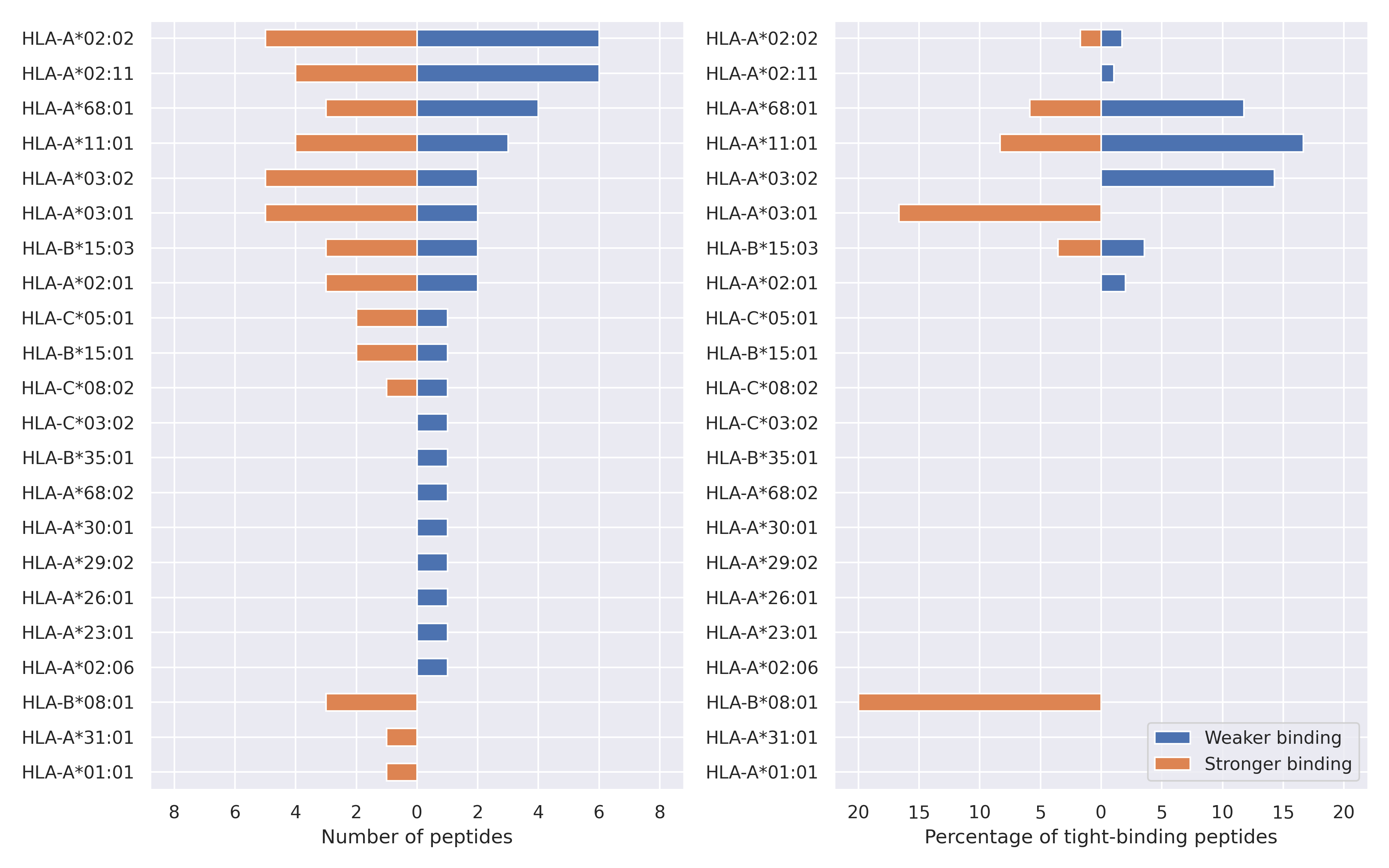
SGF 106-108 deletion
Reference FNMVYMPASWVMRIMTWLDMVDTSLSGFKLKDCVMYASAVVLLILMTARTVYD
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated FNMVYMPASWVMRIMTWLDMVDTSL‑‑‑KLKDCVMYASAVVLLILMTARTVYD
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-A*02:02 | FKLKDCVMYA | - | 8 | - |
| HLA-A*02:11 | FKLKDCVMYA | - | 12 | - |
| HLA-A*68:01 | MVDTSLSGFK | MVDTSLK | 13 | - |
| HLA-B*15:03 | FKLKDCVMY | - | 15 | - |
| HLA-A*11:01 | MVDTSLSGFK | MVDTSLK | 21 | - |
| HLA-A*11:01 | TSLSGFKLK | TSLKLK | 22 | - |
| HLA-A*03:01 | RIMTWLDMVDTSLSGFK | RIMTWLDMVDTSLK | - | 25 |
| HLA-A*02:01 | FKLKDCVMYA | - | 27 | - |
| HLA-A*68:01 | DTSLSGFKLK | DTSLKLK | 33 | - |
| HLA-A*03:02 | MVDTSLSGFK | MVDTSLK | 35 | - |
| HLA-A*68:01 | MTWLDMVDTSLSGFK | MTWLDMVDTSLK | - | 37 |
| HLA-A*03:01 | MVDTSLSGFK | MVDTSLK | 54 | - |
| HLA-A*03:02 | RIMTWLDMVDTSLSGFK | RIMTWLDMVDTSLK | - | 59 |
| HLA-C*05:01 | MVDTSLSGF | MVDTSL | 64 | - |
| HLA-A*11:01 | MTWLDMVDTSLSGFK | MTWLDMVDTSLK | - | 71 |
| HLA-A*02:06 | FKLKDCVMYA | - | 90 | - |
| HLA-A*30:01 | TSLSGFKLK | TSLKLK | 107 | - |
| HLA-A*02:02 | SGFKLKDCVMYA | - | 126 | - |
| HLA-A*11:01 | RIMTWLDMVDTSLSGFK | RIMTWLDMVDTSLK | - | 128 |
| HLA-A*03:02 | MTWLDMVDTSLSGFK | MTWLDMVDTSLK | - | 147 |
| HLA-C*08:02 | MVDTSLSGF | MVDTSL | 159 | - |
| HLA-A*03:02 | IMTWLDMVDTSLSGFK | IMTWLDMVDTSLK | - | 168 |
| HLA-A*02:11 | SGFKLKDCVMYA | - | 200 | - |
| HLA-A*02:02 | MTWLDMVDTSLSGFKL | MTWLDMVDTSLKL | - | 202 |
| HLA-A*01:01 | MVDTSLSGFKLKDCVMY | MVDTSLKLKDCVMY | - | 207 |
| HLA-C*03:02 | MVDTSLSGF | MVDTSL | 211 | - |
| HLA-A*02:02 | GFKLKDCVMYA | - | 239 | - |
| HLA-A*02:02 | TSLSGFKLKDCVMYA | TSLKLKDCVMYA | - | 244 |
| HLA-A*03:01 | TSLSGFKLK | TSLKLK | 244 | - |
| HLA-B*15:03 | MVDTSLSGF | MVDTSL | 276 | - |
| HLA-A*68:01 | TSLSGFKLK | TSLKLK | 286 | - |
| HLA-A*03:02 | TSLSGFKLK | TSLKLK | 289 | - |
| HLA-A*03:01 | IMTWLDMVDTSLSGFK | IMTWLDMVDTSLK | - | 309 |
| HLA-A*68:02 | DTSLSGFKL | DTSLKL | 313 | - |
| HLA-A*02:11 | MTWLDMVDTSLSGFKL | MTWLDMVDTSLKL | - | 346 |
| HLA-B*15:01 | DMVDTSLSGF | DMVDTSL | 362 | - |
| HLA-A*23:01 | TWLDMVDTSLSGF | - | 387 | - |
| HLA-A*02:11 | GFKLKDCVMYA | - | 394 | - |
| HLA-A*03:01 | MTWLDMVDTSLSGFK | MTWLDMVDTSLK | - | 415 |
| HLA-A*26:01 | DMVDTSLSGF | DMVDTSL | 417 | - |
| HLA-B*08:01 | MVDTSLSGFKLKDCVM | MVDTSLKLKDCVM | - | 426 |
| HLA-A*02:02 | WLDMVDTSLS | - | 441 | - |
| HLA-A*02:11 | FKLKDCVMYASAVV | - | 481 | - |
| HLA-A*11:01 | DTSLSGFKLK | DTSLKLK | 488 | - |
| HLA-B*35:01 | MVDTSLSGF | MVDTSL | 490 | - |
| HLA-A*31:01 | RIMTWLDMVDTSLSGFK | RIMTWLDMVDTSLK | - | 493 |
| HLA-B*15:03 | LSGFKLKDCVMY | LKLKDCVMY | 3466 | 22 |
| HLA-A*02:11 | LDMVDTSLSGFKL | LDMVDTSLKL | 16396 | 166 |
| HLA-A*02:02 | LDMVDTSLSGFKL | LDMVDTSLKL | 9971 | 106 |
| HLA-A*02:11 | SLSGFKLKDCV | SLKLKDCV | 122 | 9934 |
| HLA-A*02:01 | LDMVDTSLSGFKL | LDMVDTSLKL | 20579 | 311 |
| HLA-B*08:01 | SLSGFKLKDCVM | SLKLKDCVM | 929 | 36 |
| HLA-A*02:02 | SLSGFKLKDCV | SLKLKDCV | 408 | 10216 |
| HLA-C*05:01 | WLDMVDTSLSGFKL | WLDMVDTSLKL | 8375 | 417 |
| HLA-B*15:01 | SLSGFKLKDCVMY | SLKLKDCVMY | 2952 | 158 |
| HLA-B*08:01 | TSLSGFKLKDCVM | TSLKLKDCVM | 4628 | 289 |
| HLA-B*15:01 | SLSGFKLKDCVM | SLKLKDCVM | 2654 | 197 |
| HLA-A*68:01 | DMVDTSLSGFK | DMVDTSLK | 366 | 4919 |
| HLA-A*02:11 | WLDMVDTSLSGFKL | WLDMVDTSLKL | 1342 | 108 |
| HLA-C*08:02 | MVDTSLSGFKL | MVDTSLKL | 2134 | 174 |
| HLA-B*15:03 | SLSGFKLKDCVM | SLKLKDCVM | 3634 | 297 |
| HLA-A*02:01 | WLDMVDTSLSGFKL | WLDMVDTSLKL | 2214 | 195 |
| HLA-A*03:02 | WLDMVDTSLSGFK | WLDMVDTSLK | 1756 | 156 |
| HLA-A*02:02 | LSGFKLKDCVMYA | LKLKDCVMYA | 305 | 28 |
| HLA-A*02:01 | LSGFKLKDCVMYA | LKLKDCVMYA | 1788 | 165 |
| HLA-A*02:11 | LSGFKLKDCVMYA | LKLKDCVMYA | 578 | 54 |
| HLA-B*15:03 | SLSGFKLKDCVMY | SLKLKDCVMY | 3626 | 349 |
| HLA-A*03:01 | WLDMVDTSLSGFK | WLDMVDTSLK | 2900 | 287 |
| HLA-C*05:01 | MVDTSLSGFKL | MVDTSLKL | 1125 | 124 |
| HLA-A*11:01 | WLDMVDTSLSGFK | WLDMVDTSLK | 4020 | 492 |
| HLA-A*02:02 | WLDMVDTSLSGFKL | WLDMVDTSLKL | 788 | 116 |
| HLA-A*68:01 | DMVDTSLSGFKLK | DMVDTSLKLK | 1679 | 251 |
| HLA-A*02:01 | SLSGFKLKDCVMYA | SLKLKDCVMYA | 203 | 853 |
| HLA-A*02:11 | SLSGFKLKDCVMYA | SLKLKDCVMYA | 64 | 223 |
| HLA-A*02:02 | SLSGFKLKDCVMYA | SLKLKDCVMYA | 55 | 171 |
| HLA-A*29:02 | SLSGFKLKDCVMY | SLKLKDCVMY | 404 | 1192 |
| HLA-A*11:01 | MVDTSLSGFKLK | MVDTSLKLK | 143 | 50 |
| HLA-A*03:02 | MVDTSLSGFKLK | MVDTSLKLK | 312 | 127 |
| HLA-A*68:01 | MVDTSLSGFKLK | MVDTSLKLK | 602 | 246 |
| HLA-A*03:01 | MVDTSLSGFKLK | MVDTSLKLK | 441 | 190 |
NSP12 protein

P323L
Reference CVNCLDDRCILHCANFNVLFSTVFPPTSFGPLVRKIFVDGVPFVVSTGYHF
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated CVNCLDDRCILHCANFNVLFSTVFPLTSFGPLVRKIFVDGVPFVVSTGYHF
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-A*02:01 | FPPTSFGPLV | FPLTSFGPLV | 8999 | 21 |
| HLA-A*02:02 | FPPTSFGPLV | FPLTSFGPLV | 7237 | 18 |
| HLA-A*02:11 | FPPTSFGPLV | FPLTSFGPLV | 3405 | 9 |
| HLA-A*32:01 | VLFSTVFPP | VLFSTVFPL | 5415 | 27 |
| HLA-A*02:11 | LFSTVFPP | LFSTVFPL | 6706 | 35 |
| HLA-A*02:11 | ANFNVLFSTVFPP | ANFNVLFSTVFPL | 1827 | 12 |
| HLA-A*02:11 | NFNVLFSTVFPP | NFNVLFSTVFPL | 1779 | 13 |
| HLA-A*02:11 | NVLFSTVFPP | NVLFSTVFPL | 739 | 6 |
| HLA-C*03:03 | VLFSTVFPP | VLFSTVFPL | 28468 | 265 |
| HLA-C*03:04 | VLFSTVFPP | VLFSTVFPL | 28468 | 265 |
| HLA-A*02:11 | FNVLFSTVFPP | FNVLFSTVFPL | 413 | 4 |
| HLA-C*14:02 | VLFSTVFPP | VLFSTVFPL | 21407 | 220 |
| HLA-A*02:11 | FNVLFSTVFPPT | FNVLFSTVFPLT | 2618 | 27 |
| HLA-A*02:01 | LFSTVFPP | LFSTVFPL | 7475 | 81 |
| HLA-C*14:02 | LFSTVFPP | LFSTVFPL | 11979 | 138 |
| HLA-B*15:03 | VLFSTVFPP | VLFSTVFPL | 3115 | 36 |
| HLA-A*02:11 | VLFSTVFPP | VLFSTVFPL | 78 | 1 |
| HLA-C*03:02 | VLFSTVFPP | VLFSTVFPL | 20533 | 273 |
| HLA-A*02:01 | FNVLFSTVFPP | FNVLFSTVFPL | 656 | 9 |
| HLA-A*02:01 | ANFNVLFSTVFPP | ANFNVLFSTVFPL | 2569 | 37 |
| HLA-A*02:11 | FNVLFSTVFPPTS | FNVLFSTVFPLTS | 9448 | 142 |
| HLA-A*02:02 | NVLFSTVFPP | NVLFSTVFPL | 1088 | 17 |
| HLA-A*02:11 | CANFNVLFSTVFPP | CANFNVLFSTVFPL | 6143 | 96 |
| HLA-A*02:02 | FNVLFSTVFPP | FNVLFSTVFPL | 1014 | 16 |
| HLA-A*68:02 | NVLFSTVFPP | NVLFSTVFPL | 2213 | 37 |
| HLA-A*02:02 | ANFNVLFSTVFPP | ANFNVLFSTVFPL | 3126 | 53 |
| HLA-A*02:11 | PPTSFGPLV | PLTSFGPLV | 28329 | 483 |
| HLA-C*16:01 | VLFSTVFPP | VLFSTVFPL | 24733 | 432 |
| HLA-C*17:01 | VLFSTVFPP | VLFSTVFPL | 25087 | 439 |
| HLA-A*02:01 | FNVLFSTVFPPT | FNVLFSTVFPLT | 3765 | 66 |
| HLA-A*02:02 | LFSTVFPP | LFSTVFPL | 9311 | 167 |
| HLA-A*02:01 | NVLFSTVFPP | NVLFSTVFPL | 928 | 17 |
| HLA-A*02:02 | NFNVLFSTVFPP | NFNVLFSTVFPL | 2542 | 47 |
| HLA-A*02:02 | VLFSTVFPP | VLFSTVFPL | 269 | 5 |
| HLA-A*02:01 | NFNVLFSTVFPP | NFNVLFSTVFPL | 2286 | 44 |
| HLA-A*02:06 | FPPTSFGPLV | FPLTSFGPLV | 3379 | 73 |
| HLA-A*02:01 | VLFSTVFPP | VLFSTVFPL | 179 | 4 |
| HLA-A*02:06 | FNVLFSTVFPP | FNVLFSTVFPL | 1609 | 37 |
| HLA-A*02:02 | FNVLFSTVFPPT | FNVLFSTVFPLT | 4116 | 110 |
| HLA-A*02:11 | FSTVFPPTSFGPLV | FSTVFPLTSFGPLV | 6695 | 180 |
| HLA-A*02:01 | FNVLFSTVFPPTS | FNVLFSTVFPLTS | 11451 | 311 |
| HLA-A*02:06 | VLFSTVFPP | VLFSTVFPL | 368 | 10 |
| HLA-A*02:11 | ANFNVLFSTVFPPT | ANFNVLFSTVFPLT | 14639 | 409 |
| HLA-A*02:01 | FSTVFPPTSFGPLV | FSTVFPLTSFGPLV | 12596 | 425 |
| HLA-A*68:01 | PTSFGPLVR | LTSFGPLVR | 908 | 36 |
| HLA-A*02:06 | NVLFSTVFPP | NVLFSTVFPL | 1611 | 66 |
| HLA-B*15:01 | VLFSTVFPP | VLFSTVFPL | 5731 | 243 |
| HLA-B*35:01 | FPPTSFGPL | FPLTSFGPL | 304 | 13 |
| HLA-B*53:01 | FPPTSFGPL | FPLTSFGPL | 3581 | 154 |
| HLA-C*01:02 | FPPTSFGPL | FPLTSFGPL | 217 | 4855 |
| HLA-A*02:01 | CANFNVLFSTVFPP | CANFNVLFSTVFPL | 8519 | 384 |
| HLA-A*02:02 | FNVLFSTVFPPTS | FNVLFSTVFPLTS | 10920 | 497 |
| HLA-A*02:02 | FSTVFPPTSFGPLV | FSTVFPLTSFGPLV | 6958 | 322 |
| HLA-A*02:11 | VFPPTSFGPLV | VFPLTSFGPLV | 4242 | 199 |
| HLA-A*02:02 | CANFNVLFSTVFPP | CANFNVLFSTVFPL | 8183 | 391 |
| HLA-A*02:11 | NFNVLFSTVFPPT | NFNVLFSTVFPLT | 9111 | 472 |
| HLA-A*02:06 | ANFNVLFSTVFPP | ANFNVLFSTVFPL | 5191 | 304 |
| HLA-A*02:06 | NFNVLFSTVFPP | NFNVLFSTVFPL | 4220 | 274 |
| HLA-A*02:06 | FNVLFSTVFPPT | FNVLFSTVFPLT | 4868 | 388 |
| HLA-B*35:03 | FPPTSFGPL | FPLTSFGPL | 286 | 24 |
| HLA-A*03:01 | PTSFGPLVRK | LTSFGPLVRK | 554 | 47 |
| HLA-A*31:01 | PTSFGPLVR | LTSFGPLVR | 4402 | 390 |
| HLA-A*11:01 | PTSFGPLVR | LTSFGPLVR | 4068 | 379 |
| HLA-A*03:02 | PTSFGPLVRK | LTSFGPLVRK | 868 | 88 |
| HLA-B*07:02 | STVFPPTSFGPL | STVFPLTSFGPL | 777 | 79 |
| HLA-A*68:01 | PTSFGPLVRK | LTSFGPLVRK | 308 | 34 |
| HLA-B*07:02 | TVFPPTSFGPL | TVFPLTSFGPL | 1498 | 171 |
| HLA-A*02:11 | STVFPPTSFGPLV | STVFPLTSFGPLV | 2427 | 285 |
| HLA-A*02:11 | NVLFSTVFPPT | NVLFSTVFPLT | 1183 | 139 |
| HLA-A*33:03 | PTSFGPLVR | LTSFGPLVR | 3228 | 381 |
| HLA-A*26:01 | STVFPPTSF | STVFPLTSF | 786 | 95 |
| HLA-B*07:02 | VFPPTSFGPL | VFPLTSFGPL | 339 | 43 |
| HLA-A*02:01 | NVLFSTVFPPT | NVLFSTVFPLT | 2111 | 290 |
| HLA-B*42:01 | FSTVFPPTSFGPL | FSTVFPLTSFGPL | 1320 | 185 |
| HLA-B*42:01 | STVFPPTSFGPL | STVFPLTSFGPL | 738 | 111 |
| HLA-A*02:01 | VLFSTVFPPTSF | VLFSTVFPLTSF | 2225 | 342 |
| HLA-B*08:01 | FPPTSFGPL | FPLTSFGPL | 973 | 153 |
| HLA-B*42:01 | TVFPPTSFGPL | TVFPLTSFGPL | 843 | 142 |
| HLA-A*11:01 | PTSFGPLVRK | LTSFGPLVRK | 123 | 21 |
| HLA-B*42:01 | VFPPTSFGPL | VFPLTSFGPL | 274 | 47 |
| HLA-A*02:01 | VLFSTVFPPTS | VLFSTVFPLTS | 708 | 124 |
| HLA-B*07:02 | FSTVFPPTSFGPL | FSTVFPLTSFGPL | 2537 | 466 |
| HLA-B*07:02 | FPPTSFGPL | FPLTSFGPL | 42 | 8 |
| HLA-A*02:11 | VLFSTVFPPTSF | VLFSTVFPLTSF | 1019 | 195 |
| HLA-A*25:01 | STVFPPTSF | STVFPLTSF | 1533 | 296 |
| HLA-B*07:02 | FPPTSFGPLV | FPLTSFGPLV | 1131 | 236 |
| HLA-B*42:01 | FPPTSFGPL | FPLTSFGPL | 21 | 5 |
| HLA-A*26:01 | FSTVFPPTSF | FSTVFPLTSF | 1667 | 398 |
| HLA-B*51:01 | FPPTSFGPL | FPLTSFGPL | 1809 | 433 |
| HLA-A*02:11 | VLFSTVFPPTS | VLFSTVFPLTS | 309 | 74 |
| HLA-B*42:01 | FPPTSFGPLV | FPLTSFGPLV | 368 | 89 |
| HLA-A*02:02 | VLFSTVFPPTS | VLFSTVFPLTS | 1210 | 377 |
| HLA-C*03:02 | FPPTSFGPL | FPLTSFGPL | 960 | 313 |
| HLA-A*32:01 | STVFPPTSF | STVFPLTSF | 230 | 86 |
| HLA-B*58:01 | FSTVFPPTSF | FSTVFPLTSF | 88 | 33 |
| HLA-B*15:01 | STVFPPTSF | STVFPLTSF | 130 | 49 |
| HLA-A*02:01 | VLFSTVFPPT | VLFSTVFPLT | 61 | 23 |
| HLA-C*03:03 | FPPTSFGPL | FPLTSFGPL | 375 | 145 |
| HLA-C*03:04 | FPPTSFGPL | FPLTSFGPL | 375 | 145 |
| HLA-A*02:02 | VLFSTVFPPT | VLFSTVFPLT | 113 | 44 |
| HLA-C*16:01 | FSTVFPPTS | FSTVFPLTS | 1127 | 460 |
| HLA-C*14:02 | LFSTVFPPTSF | LFSTVFPLTSF | 396 | 950 |
| HLA-C*01:02 | VFPPTSFGPL | VFPLTSFGPL | 429 | 997 |
| HLA-A*02:02 | TVFPPTSFGPLV | TVFPLTSFGPLV | 789 | 343 |
| HLA-A*68:02 | STVFPPTSFGPLV | STVFPLTSFGPLV | 169 | 74 |
| HLA-A*02:11 | VLFSTVFPPT | VLFSTVFPLT | 25 | 11 |
| HLA-A*03:01 | TVFPPTSFGPLVRK | TVFPLTSFGPLVRK | 92 | 198 |
| HLA-C*14:02 | VFPPTSFGPL | VFPLTSFGPL | 47 | 99 |
| HLA-A*23:01 | LFSTVFPPTSF | LFSTVFPLTSF | 370 | 179 |
NSP13 protein
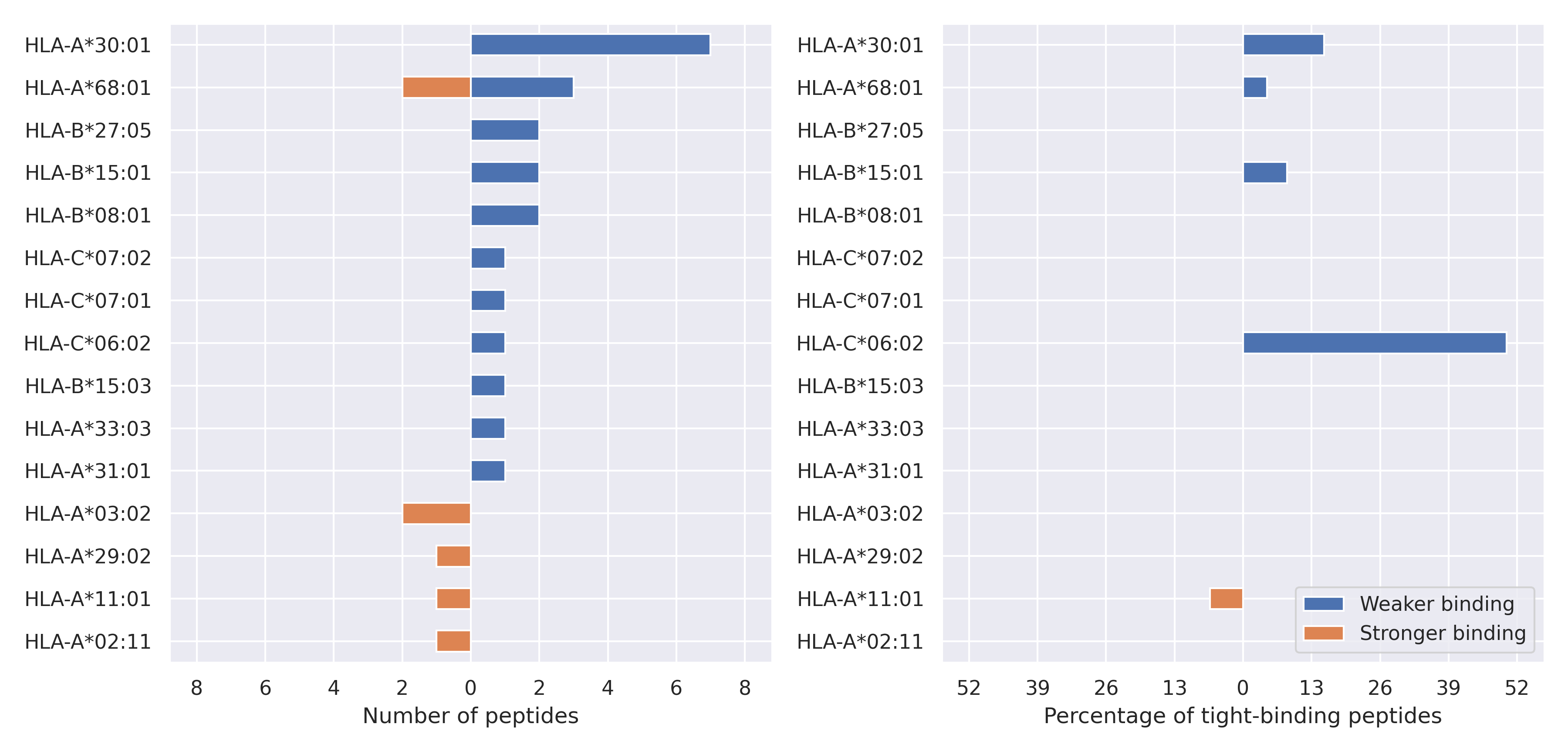
R392C
Reference TADIVVFDEISMATNYDLSVVNARLRAKHYVYIGDPAQLPAPRTLLTKGTL
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated TADIVVFDEISMATNYDLSVVNARLCAKHYVYIGDPAQLPAPRTLLTKGTL
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-A*68:01 | DLSVVNARLR | DLSVVNARLC | 44 | 30873 |
| HLA-C*06:02 | LRAKHYVYI | LCAKHYVYI | 36 | 16159 |
| HLA-A*68:01 | LSVVNARLR | LSVVNARLC | 140 | 32767 |
| HLA-A*33:03 | DLSVVNARLR | DLSVVNARLC | 154 | 32767 |
| HLA-A*31:01 | LSVVNARLR | LSVVNARLC | 261 | 30993 |
| HLA-C*07:01 | LRAKHYVYI | LCAKHYVYI | 133 | 15165 |
| HLA-B*27:05 | LRAKHYVYI | LCAKHYVYI | 379 | 32767 |
| HLA-A*30:01 | RLRAKHYVY | RLCAKHYVY | 27 | 1483 |
| HLA-B*27:05 | RLRAKHYVYI | RLCAKHYVYI | 208 | 10096 |
| HLA-C*07:02 | LRAKHYVYI | LCAKHYVYI | 433 | 17614 |
| HLA-A*30:01 | RAKHYVYIG | CAKHYVYIG | 76 | 2630 |
| HLA-A*68:01 | TNYDLSVVNARLR | TNYDLSVVNARLC | 477 | 15989 |
| HLA-A*30:01 | RLRAKHYV | RLCAKHYV | 218 | 5721 |
| HLA-A*30:01 | RLRAKHYVYI | RLCAKHYVYI | 17 | 427 |
| HLA-A*30:01 | RAKHYVYIGDPA | CAKHYVYIGDPA | 188 | 4601 |
| HLA-A*30:01 | RAKHYVYI | CAKHYVYI | 211 | 4134 |
| HLA-B*08:01 | RLRAKHYV | RLCAKHYV | 363 | 6671 |
| HLA-A*30:01 | ARLRAKHYVYI | ARLCAKHYVYI | 422 | 6081 |
| HLA-B*08:01 | RLRAKHYVYI | RLCAKHYVYI | 230 | 2547 |
| HLA-B*15:03 | LRAKHYVY | LCAKHYVY | 395 | 4119 |
| HLA-A*29:02 | RLRAKHYVY | RLCAKHYVY | 535 | 101 |
| HLA-A*02:11 | RLRAKHYV | RLCAKHYV | 1483 | 352 |
| HLA-A*68:01 | VVNARLRAK | VVNARLCAK | 1428 | 381 |
| HLA-A*11:01 | VVNARLRAK | VVNARLCAK | 145 | 44 |
| HLA-A*03:02 | VVNARLRAK | VVNARLCAK | 257 | 79 |
| HLA-B*15:01 | ARLRAKHYVY | ARLCAKHYVY | 402 | 1223 |
| HLA-B*15:01 | RLRAKHYVY | RLCAKHYVY | 49 | 133 |
| HLA-A*68:01 | SVVNARLRAK | SVVNARLCAK | 158 | 63 |
| HLA-A*03:02 | SVVNARLRAK | SVVNARLCAK | 196 | 86 |
NSP14 protein
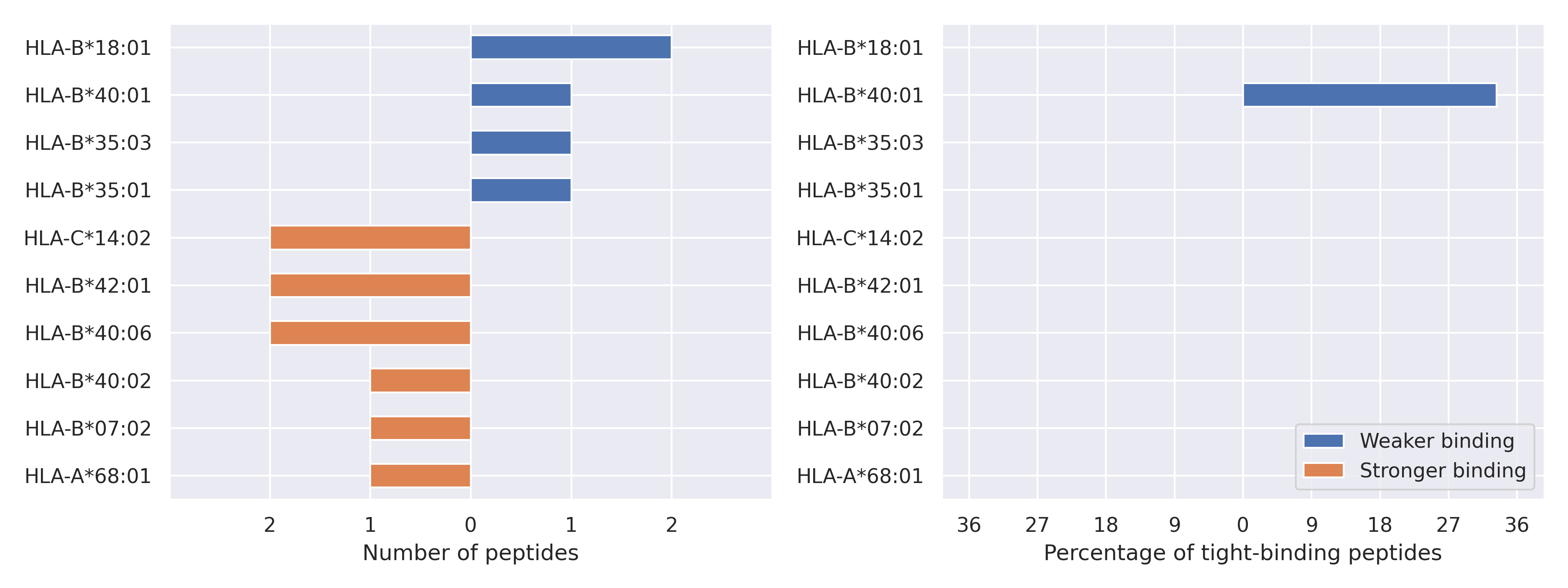
I42V
Reference GLHPTQAPTHLSVDTKFKTEGLCVDIPGIPKDMTYRRLISMMGFKMNYQVN
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated GLHPTQAPTHLSVDTKFKTEGLCVDVPGIPKDMTYRRLISMMGFKMNYQVN
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-A*68:01 | DIPGIPKDMTYRR | DVPGIPKDMTYRR | 570 | 271 |
D301G
Reference MTRCLAVHECFVKRVDWTIEYPIIGDELKINAACRKVQHMVVKAALLADKF
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated MTRCLAVHECFVKRVDWTIEYPIIGGELKINAACRKVQHMVVKAALLADKF
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-B*18:01 | DELKINAA | GELKINAA | 365 | 11741 |
| HLA-B*40:02 | DELKINAAC | GELKINAAC | 4412 | 153 |
| HLA-B*18:01 | DELKINAAC | GELKINAAC | 435 | 11957 |
| HLA-B*40:06 | DELKINAA | GELKINAA | 7148 | 373 |
| HLA-B*40:06 | DELKINAAC | GELKINAAC | 5446 | 438 |
| HLA-B*07:02 | YPIIGDEL | YPIIGGEL | 1141 | 185 |
| HLA-B*42:01 | YPIIGDEL | YPIIGGEL | 288 | 70 |
| HLA-B*42:01 | YPIIGDELKI | YPIIGGELKI | 1320 | 359 |
| HLA-C*14:02 | EYPIIGDEL | EYPIIGGEL | 678 | 236 |
| HLA-C*14:02 | IEYPIIGDEL | IEYPIIGGEL | 796 | 293 |
| HLA-B*35:03 | YPIIGDEL | YPIIGGEL | 345 | 853 |
| HLA-B*35:01 | YPIIGDEL | YPIIGGEL | 364 | 758 |
| HLA-B*40:01 | IEYPIIGDEL | IEYPIIGGEL | 49 | 99 |
NSP15 protein
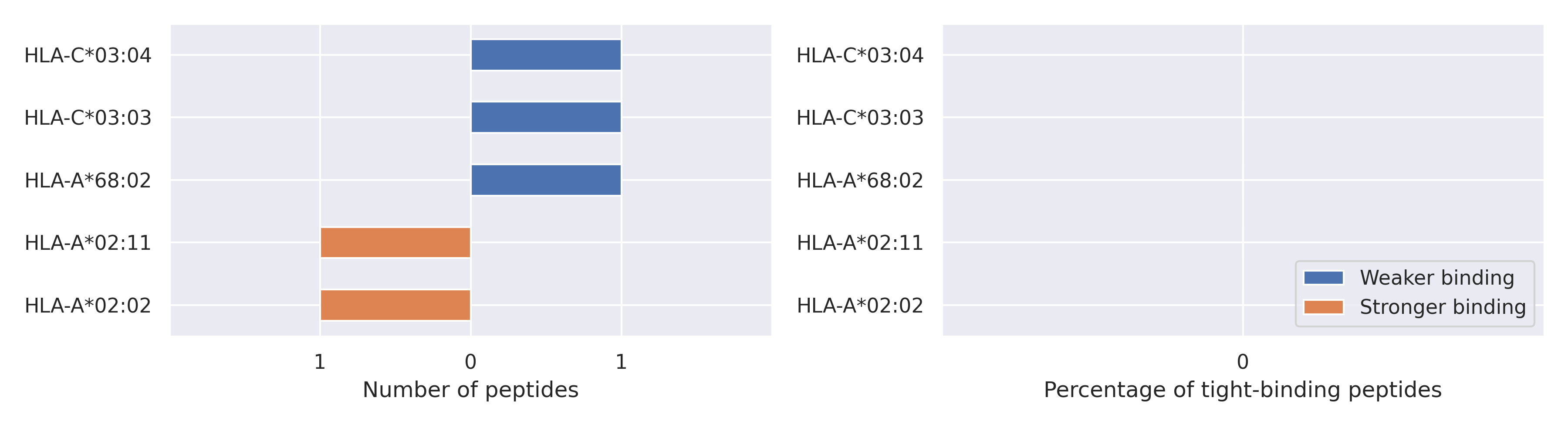
T112I
Reference DYKRDAPAHISTIGVCSMTDIAKKPTETICAPLTVFFDGRVDGQVDLFRNA
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated DYKRDAPAHISTIGVCSMTDIAKKPIETICAPLTVFFDGRVDGQVDLFRNA
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-A*02:02 | SMTDIAKKPT | SMTDIAKKPI | 3353 | 375 |
| HLA-A*68:02 | TETICAPLTV | IETICAPLTV | 64 | 161 |
| HLA-C*03:03 | IAKKPTETI | IAKKPIETI | 497 | 1187 |
| HLA-C*03:04 | IAKKPTETI | IAKKPIETI | 497 | 1187 |
| HLA-A*02:11 | SMTDIAKKPTETI | SMTDIAKKPIETI | 560 | 263 |