Summary
This strain contains 70 protein-level mutations:
- 36 mutations in Spike protein: T19I, L24S, A27S, V83A, G142D, H146Q, E180V, Q183E, V213E, G339H, R346T, L368I, S371F, S373P, 375-376 ST->FA, D405N, R408S, K417N, N440K, 445-446 VG->PS, N460K, 477-478 ST->NR, E484A, F486P, F490S, Q498R, N501Y, Y505H, D614G, H655Y, N679K, P681H, N764K, D796Y, Q954H, N969K
- 4 mutations in N protein: P13L, S33G, 203-204 RG->KR, S413R
- 2 mutations in M protein: Q19E, A63T
- 2 mutations in E protein: T9I, T11A
- 1 mutation in NS3 protein: T223I
- 1 mutation in NS8 protein: G8stop
- 4 mutations in NS9b protein: I5T, P10S, E27V, N55S
- 1 mutation in NS9c protein: G50N
- 2 mutations in NSP1 protein: K47R, S135R
- 2 mutations in NSP3 protein: T24I, G489S
- 4 mutations in NSP4 protein: L264F, T327I, L438F, T492I
- 1 mutation in NSP5 protein: P132H
- 2 mutations in NSP6 protein: 106-108 SGF->TSL, L260F
- 2 mutations in NSP12 protein: P323L, G671S
- 2 mutations in NSP13 protein: S36P, R392C
- 3 mutations in NSP14 protein: I42V, L157F, D222Y
- 1 mutation in NSP15 protein: T112I
We identified all possible linear viral peptides affected by these mutations. Whenever it was possible, we matched the reference peptide with the mutated one. For example, D -> L mutation transformed SDNGPQNQR to SLNGPQNQR. Cases when it was not meaningful included deletions and insertions at the flanks of the peptide, e.g., HV deletion in NVTWFHAIHV peptide.
Then, we predicted binding affinities between the selected peptides and frequent HLA alleles. Predictions were made with NetMHCpan-4.1 and NetMHCIIpan-4.0. The binding affinities were classifies into three groups:
- Tight binding (IC50 affinity ≤ 50 nM)
- Moderate binding (50 nM < IC50 affinity ≤ 500 nM)
- Weak/no binding (IC50 affinity > 500 nM)
Here we report HLA-peptide interactions whose affinity was altered by at least two folds. Note that mutations with empty set of altered interactions are not showed.
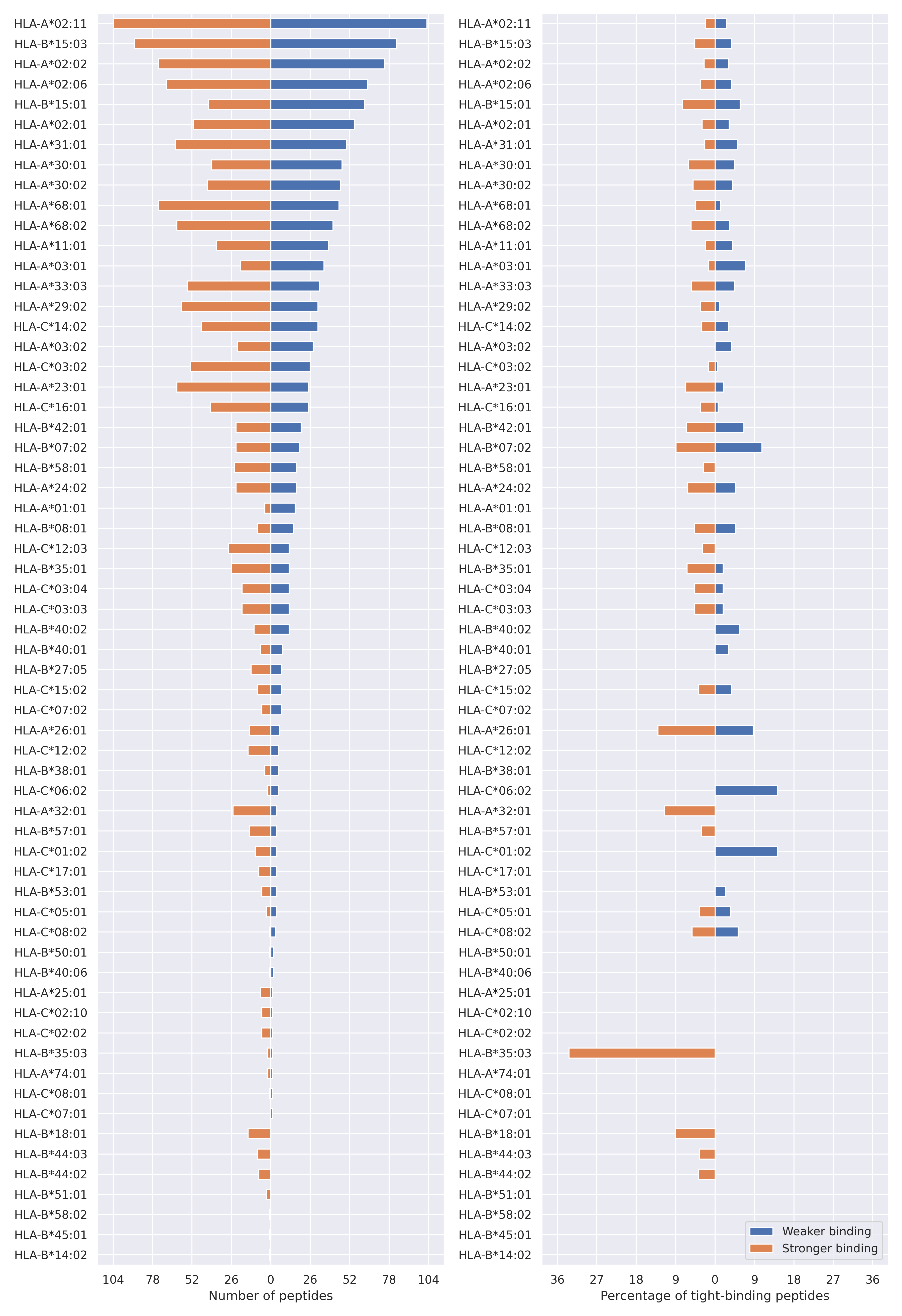
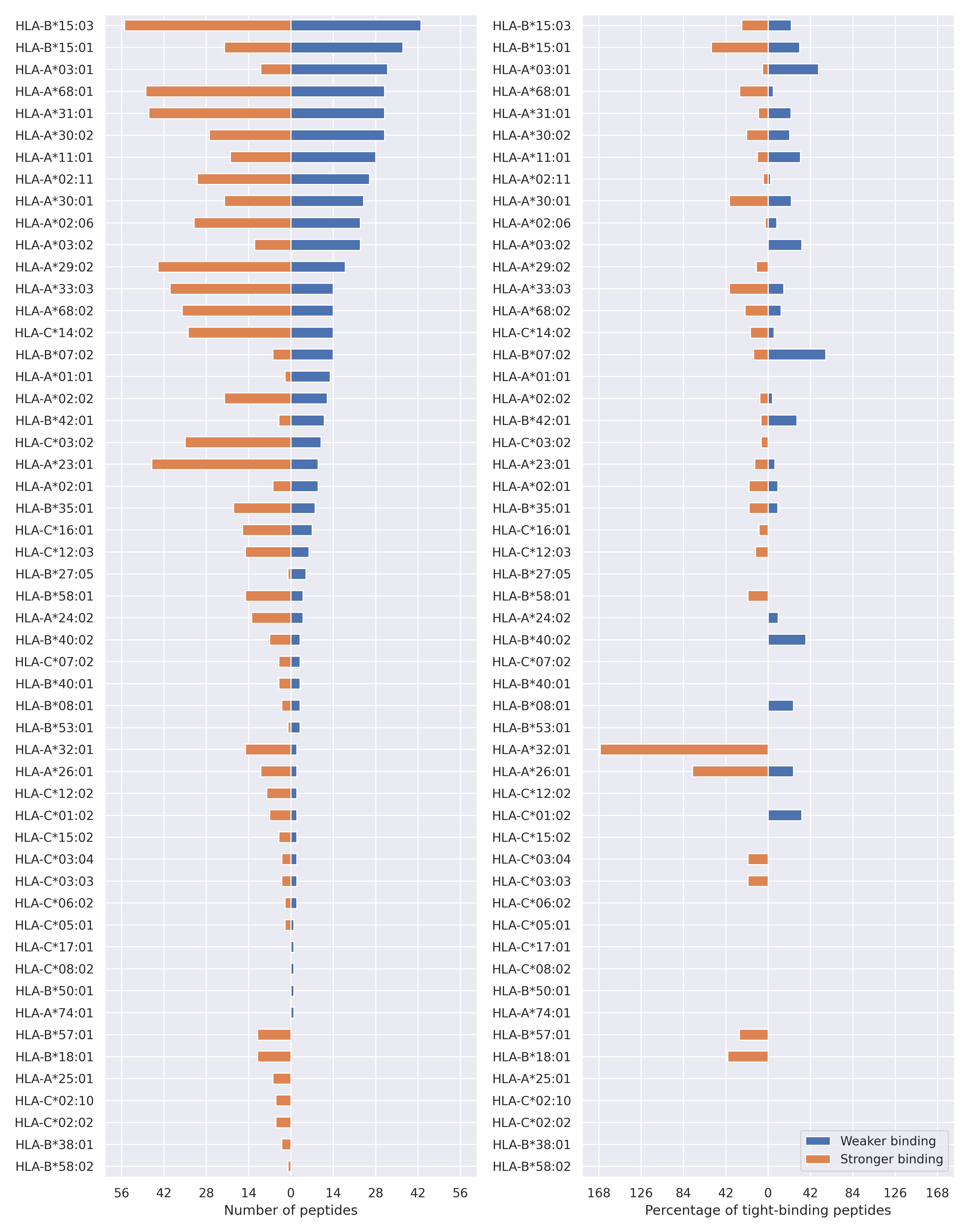
The total number of HLA-peptide interactions affected by the mutations
For each allele we calculated the number of peptides with increased and decreased affinitiy (two or more times). The absolute number of such peptides is showed on the left side of the figure (blue bars — weaker binding, orange bars — stronger).
While absolute numbers are of interest, they do not reflect the initial number of tighly binding peptides for the specific allele. Roughly speaking, if an allele had high number of tight binders, then vanished affinity of several peptides will not affect much the integral ability of peptide presentation. To the contrary, low number of vanished peptides could be important if the particular allele initially had narrow epitope repertoire. To account for this effect, we normalized the absolute numbers of new/vanished tight binders (IC50 affinity ≤ 50 nM) by the total number of tight binders for each allele. To avoid possible divisions by zero, we used + 1 regularization term in all denominators. For example, if some allele had no tight binders in the reference genome, and three new tight binders appeared as a result of the mutation, the relative increase will be 300%. The results are showed in the right part of the figure. Protein-specific plots are listed below.
Spike protein
T19I
Reference MFVFLVLLPLVSSQCVNLTTRTQLPPAYTNSFTRGVYYPDKVFR
|||||||||||||||||| |||||||||||||||||||||||||
Mutated MFVFLVLLPLVSSQCVNLITRTQSPPSYTNSFTRGVYYPDKVFR
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-B*58:01 | VSSQCVNLT | VSSQCVNLI | 3986 | 159 |
| HLA-C*15:02 | VSSQCVNLT | VSSQCVNLI | 10208 | 479 |
| HLA-A*02:02 | LVSSQCVNLT | LVSSQCVNLI | 3376 | 271 |
| HLA-A*02:11 | VLLPLVSSQCVNLT | VLLPLVSSQCVNLI | 4460 | 394 |
| HLA-A*30:01 | VNLTTRTQLPPA | VNLITRTQSPPS | 254 | 1670 |
| HLA-A*30:01 | LTTRTQLPPA | LITRTQSPPS | 82 | 460 |
| HLA-A*30:01 | TTRTQLPPA | ITRTQSPPS | 9 | 33 |
| HLA-B*15:03 | TTRTQLPPAY | ITRTQSPPSY | 888 | 396 |
L24S
Reference MFVFLVLLPLVSSQCVNLTTRTQLPPAYTNSFTRGVYYPDKVFRSSVLH
||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated MFVFLVLLPLVSSQCVNLITRTQSPPSYTNSFTRGVYYPDKVFRSSVLH
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-A*11:01 | LPPAYTNSFTR | SPPSYTNSFTR | 3363 | 229 |
| HLA-A*30:01 | VNLTTRTQLPPA | VNLITRTQSPPS | 254 | 1670 |
| HLA-B*15:01 | QLPPAYTNSF | QSPPSYTNSF | 300 | 1776 |
| HLA-A*30:01 | LTTRTQLPPA | LITRTQSPPS | 82 | 460 |
| HLA-B*53:01 | LPPAYTNSF | SPPSYTNSF | 366 | 1877 |
| HLA-A*31:01 | LPPAYTNSFTR | SPPSYTNSFTR | 757 | 195 |
| HLA-B*35:01 | LPPAYTNSF | SPPSYTNSF | 43 | 161 |
| HLA-A*30:01 | TTRTQLPPA | ITRTQSPPS | 9 | 33 |
| HLA-B*15:03 | TTRTQLPPAY | ITRTQSPPSY | 888 | 396 |
| HLA-B*07:02 | LPPAYTNSF | SPPSYTNSF | 293 | 137 |
A27S
Reference FVFLVLLPLVSSQCVNLTTRTQLPPAYTNSFTRGVYYPDKVFRSSVLHSTQ
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated FVFLVLLPLVSSQCVNLITRTQSPPSYTNSFTRGVYYPDKVFRSSVLHSTQ
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-A*11:01 | LPPAYTNSFTR | SPPSYTNSFTR | 3363 | 229 |
| HLA-A*30:01 | VNLTTRTQLPPA | VNLITRTQSPPS | 254 | 1670 |
| HLA-B*15:01 | QLPPAYTNSF | QSPPSYTNSF | 300 | 1776 |
| HLA-A*30:01 | LTTRTQLPPA | LITRTQSPPS | 82 | 460 |
| HLA-B*53:01 | LPPAYTNSF | SPPSYTNSF | 366 | 1877 |
| HLA-A*31:01 | LPPAYTNSFTR | SPPSYTNSFTR | 757 | 195 |
| HLA-B*35:01 | LPPAYTNSF | SPPSYTNSF | 43 | 161 |
| HLA-A*30:01 | TTRTQLPPA | ITRTQSPPS | 9 | 33 |
| HLA-A*68:02 | AYTNSFTRGV | SYTNSFTRGV | 350 | 114 |
| HLA-C*14:02 | AYTNSFTRGVYY | SYTNSFTRGVYY | 444 | 145 |
| HLA-C*14:02 | AYTNSFTRGVY | SYTNSFTRGVY | 431 | 145 |
| HLA-B*15:03 | TTRTQLPPAY | ITRTQSPPSY | 888 | 396 |
| HLA-B*07:02 | LPPAYTNSF | SPPSYTNSF | 293 | 137 |
V83A
Reference FFSNVTWFHAIHVSGTNGTKRFDNPVLPFNDGVYFASTEKSNIIRGWIFGT
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated FFSNVTWFHAIHVSGTNGTKRFDNPALPFNDGVYFASTEKSNIIRGWIFGT
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-A*02:11 | FDNPVLPFNDGV | FDNPALPFNDGV | 198 | 2056 |
| HLA-A*68:02 | NGTKRFDNPV | NGTKRFDNPA | 249 | 2355 |
| HLA-A*02:01 | FDNPVLPFNDGV | FDNPALPFNDGV | 426 | 3508 |
| HLA-A*02:02 | FDNPVLPFNDGV | FDNPALPFNDGV | 257 | 1936 |
| HLA-A*02:06 | FDNPVLPFNDGV | FDNPALPFNDGV | 55 | 374 |
| HLA-A*02:06 | NPVLPFNDGV | NPALPFNDGV | 298 | 1704 |
| HLA-A*68:02 | NPVLPFNDGV | NPALPFNDGV | 97 | 263 |
G142D
Reference LLIVNNATNVVIKVCEFQFCNDPFLGVYYHKNNKSWMESEFRVYSSANNCT
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated LLIVNNATNVVIKVCEFQFCNDPFLDVYYQKNNKSWMESEFRVYSSANNCT
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-A*03:01 | GVYYHKNNK | DVYYQKNNK | 27 | 4630 |
| HLA-A*30:01 | GVYYHKNNK | DVYYQKNNK | 51 | 7391 |
| HLA-A*11:01 | GVYYHKNNK | DVYYQKNNK | 51 | 2839 |
| HLA-A*03:02 | GVYYHKNNK | DVYYQKNNK | 119 | 5703 |
| HLA-A*31:01 | GVYYHKNNK | DVYYQKNNK | 375 | 9562 |
| HLA-A*68:02 | FCNDPFLGV | FCNDPFLDV | 281 | 3555 |
| HLA-A*68:01 | GVYYHKNNK | DVYYQKNNK | 363 | 33 |
| HLA-A*02:06 | FCNDPFLGV | FCNDPFLDV | 116 | 1132 |
| HLA-A*02:11 | FCNDPFLGV | FCNDPFLDV | 87 | 751 |
| HLA-A*02:06 | EFQFCNDPFLGV | EFQFCNDPFLDV | 87 | 554 |
| HLA-A*30:02 | CNDPFLGVY | CNDPFLDVY | 310 | 1959 |
| HLA-A*02:02 | FCNDPFLGV | FCNDPFLDV | 365 | 2187 |
| HLA-A*02:06 | FQFCNDPFLGVY | FQFCNDPFLDVY | 135 | 784 |
| HLA-A*30:02 | FCNDPFLGVY | FCNDPFLDVY | 408 | 1738 |
| HLA-A*02:11 | EFQFCNDPFLGV | EFQFCNDPFLDV | 334 | 1214 |
| HLA-A*02:02 | EFQFCNDPFLGV | EFQFCNDPFLDV | 477 | 1725 |
| HLA-A*02:11 | FQFCNDPFLGVY | FQFCNDPFLDVY | 488 | 1737 |
| HLA-A*02:06 | FQFCNDPFLGV | FQFCNDPFLDV | 7 | 21 |
| HLA-A*02:11 | FQFCNDPFLGV | FQFCNDPFLDV | 17 | 49 |
| HLA-A*02:01 | FQFCNDPFLGV | FQFCNDPFLDV | 39 | 111 |
| HLA-A*30:02 | FQFCNDPFLGVY | FQFCNDPFLDVY | 336 | 923 |
| HLA-A*02:02 | FQFCNDPFLGV | FQFCNDPFLDV | 45 | 122 |
| HLA-B*15:01 | FQFCNDPFLGVY | FQFCNDPFLDVY | 28 | 56 |
H146Q
Reference NNATNVVIKVCEFQFCNDPFLGVYYHKNNKSWMESEFRVYSSANNCTFEYV
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated NNATNVVIKVCEFQFCNDPFLDVYYQKNNKSWMESEFRVYSSANNCTFEYV
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-A*03:01 | GVYYHKNNK | DVYYQKNNK | 27 | 4630 |
| HLA-A*30:01 | GVYYHKNNK | DVYYQKNNK | 51 | 7391 |
| HLA-B*15:01 | YHKNNKSWMESEF | YQKNNKSWMESEF | 15739 | 226 |
| HLA-B*15:01 | YHKNNKSWM | YQKNNKSWM | 18557 | 271 |
| HLA-A*11:01 | GVYYHKNNK | DVYYQKNNK | 51 | 2839 |
| HLA-A*03:02 | GVYYHKNNK | DVYYQKNNK | 119 | 5703 |
| HLA-A*31:01 | GVYYHKNNK | DVYYQKNNK | 375 | 9562 |
| HLA-B*15:03 | YHKNNKSWM | YQKNNKSWM | 1065 | 50 |
| HLA-A*68:01 | GVYYHKNNK | DVYYQKNNK | 363 | 33 |
| HLA-B*15:03 | YHKNNKSWMESEF | YQKNNKSWMESEF | 1283 | 143 |
| HLA-A*24:02 | YYHKNNKSW | YYQKNNKSW | 116 | 58 |
E180V
Reference SEFRVYSSANNCTFEYVSQPFLMDLEGKQGNFKNLREFVFKNIDGYFKIYS
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated SEFRVYSSANNCTFEYVSQPFLMDLVGKEGNFKNLREFVFKNIDGYFKIYS
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-A*02:06 | SQPFLMDLE | SQPFLMDLV | 11827 | 43 |
| HLA-A*02:11 | SQPFLMDLE | SQPFLMDLV | 25287 | 173 |
| HLA-A*02:02 | SQPFLMDLE | SQPFLMDLV | 16774 | 115 |
| HLA-A*11:01 | LEGKQGNFK | LVGKEGNFK | 19398 | 215 |
| HLA-A*68:02 | YVSQPFLMDLE | YVSQPFLMDLV | 21186 | 274 |
| HLA-A*02:06 | VSQPFLMDLE | VSQPFLMDLV | 15579 | 288 |
| HLA-A*03:02 | LEGKQGNFK | LVGKEGNFK | 21955 | 465 |
| HLA-C*15:02 | VSQPFLMDLE | VSQPFLMDLV | 13114 | 340 |
| HLA-A*02:11 | YVSQPFLMDLE | YVSQPFLMDLV | 9341 | 419 |
| HLA-A*02:06 | YVSQPFLMDLE | YVSQPFLMDLV | 4972 | 233 |
| HLA-A*02:02 | YVSQPFLMDLE | YVSQPFLMDLV | 5038 | 261 |
| HLA-A*02:11 | FLMDLEGKQ | FLMDLVGKE | 1662 | 133 |
| HLA-A*02:11 | FLMDLEGKQG | FLMDLVGKEG | 3366 | 340 |
| HLA-A*02:02 | FLMDLEGKQ | FLMDLVGKE | 1621 | 220 |
| HLA-A*68:01 | DLEGKQGNFK | DLVGKEGNFK | 2888 | 402 |
| HLA-A*02:02 | FLMDLEGKQG | FLMDLVGKEG | 561 | 86 |
Q183E
Reference RVYSSANNCTFEYVSQPFLMDLEGKQGNFKNLREFVFKNIDGYFKIYSKHT
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated RVYSSANNCTFEYVSQPFLMDLVGKEGNFKNLREFVFKNIDGYFKIYSKHT
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-A*31:01 | KQGNFKNLR | KEGNFKNLR | 17 | 1692 |
| HLA-A*11:01 | LEGKQGNFK | LVGKEGNFK | 19398 | 215 |
| HLA-A*03:02 | LEGKQGNFK | LVGKEGNFK | 21955 | 465 |
| HLA-B*15:03 | KQGNFKNLREF | KEGNFKNLREF | 42 | 1050 |
| HLA-A*31:01 | GKQGNFKNLR | GKEGNFKNLR | 475 | 9702 |
| HLA-A*02:11 | FLMDLEGKQ | FLMDLVGKE | 1662 | 133 |
| HLA-A*02:11 | FLMDLEGKQG | FLMDLVGKEG | 3366 | 340 |
| HLA-B*15:03 | KQGNFKNLREFVF | KEGNFKNLREFVF | 323 | 2837 |
| HLA-A*02:02 | FLMDLEGKQ | FLMDLVGKE | 1621 | 220 |
| HLA-A*68:01 | DLEGKQGNFK | DLVGKEGNFK | 2888 | 402 |
| HLA-A*02:02 | FLMDLEGKQG | FLMDLVGKEG | 561 | 86 |
V213E
Reference NLREFVFKNIDGYFKIYSKHTPINLVRDLPQGFSALEPLVDLPIGINITRF
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated NLREFVFKNIDGYFKIYSKHTPINLERDLPQGFSALEPLVDLPIGINITRF
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-A*68:02 | YSKHTPINLV | YSKHTPINLE | 199 | 19799 |
| HLA-B*18:01 | LVRDLPQGF | LERDLPQGF | 22292 | 292 |
| HLA-C*03:02 | LVRDLPQGF | LERDLPQGF | 259 | 18781 |
| HLA-A*02:11 | KIYSKHTPINLV | KIYSKHTPINLE | 104 | 7199 |
| HLA-B*40:02 | LVRDLPQGFSAL | LERDLPQGFSAL | 17495 | 450 |
| HLA-B*15:01 | LVRDLPQGF | LERDLPQGF | 160 | 2582 |
| HLA-C*01:02 | VRDLPQGFSAL | ERDLPQGFSAL | 431 | 4216 |
| HLA-B*42:01 | TPINLVRDL | TPINLERDL | 43 | 173 |
| HLA-B*42:01 | HTPINLVRDL | HTPINLERDL | 250 | 983 |
| HLA-B*07:02 | TPINLVRDL | TPINLERDL | 213 | 783 |
G339H
Reference QTSNFRVQPTESIVRFPNITNLCPFGEVFNATRFASVYAWNRKRISNCVAD
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated QTSNFRVQPTESIVRFPNITNLCPFHEVFNATTFASVYAWNRKRISNCVAD
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-B*38:01 | FGEVFNATRF | FHEVFNATTF | 16707 | 108 |
| HLA-A*68:01 | FGEVFNATR | FHEVFNATT | 338 | 32767 |
| HLA-A*68:01 | CPFGEVFNATR | CPFHEVFNATT | 457 | 32129 |
| HLA-B*18:01 | GEVFNATRF | HEVFNATTF | 971 | 24 |
| HLA-A*68:02 | GEVFNATRFA | HEVFNATTFA | 879 | 50 |
| HLA-B*15:03 | GEVFNATRF | HEVFNATTF | 359 | 26 |
| HLA-B*15:03 | FGEVFNATRF | FHEVFNATTF | 694 | 60 |
| HLA-A*02:11 | NLCPFGEV | NLCPFHEV | 2369 | 286 |
| HLA-B*18:01 | GEVFNATRFASVY | HEVFNATTFASVY | 2165 | 265 |
| HLA-A*02:02 | FGEVFNATRFA | FHEVFNATTFA | 2678 | 413 |
| HLA-B*18:01 | FGEVFNATRF | FHEVFNATTF | 1074 | 179 |
| HLA-A*02:06 | FGEVFNATRFA | FHEVFNATTFA | 1063 | 209 |
| HLA-B*40:01 | FGEVFNATRF | FHEVFNATTF | 1994 | 435 |
| HLA-B*40:02 | FGEVFNATRF | FHEVFNATTF | 1615 | 380 |
| HLA-A*68:02 | FGEVFNATRFA | FHEVFNATTFA | 939 | 230 |
| HLA-A*02:11 | NITNLCPFGEV | NITNLCPFHEV | 1609 | 422 |
| HLA-B*35:01 | CPFGEVFNATRF | CPFHEVFNATTF | 1090 | 291 |
| HLA-A*02:06 | ITNLCPFGEV | ITNLCPFHEV | 324 | 104 |
| HLA-B*40:02 | GEVFNATRF | HEVFNATTF | 283 | 96 |
| HLA-A*02:11 | ITNLCPFGEV | ITNLCPFHEV | 412 | 147 |
| HLA-A*02:02 | NLCPFGEVFNA | NLCPFHEVFNA | 182 | 472 |
| HLA-B*15:03 | GEVFNATRFASVY | HEVFNATTFASVY | 1058 | 423 |
| HLA-A*23:01 | PFGEVFNATRF | PFHEVFNATTF | 1002 | 401 |
| HLA-A*68:02 | TNLCPFGEV | TNLCPFHEV | 963 | 398 |
| HLA-A*02:01 | NLCPFGEVFNA | NLCPFHEVFNA | 164 | 354 |
| HLA-B*40:01 | GEVFNATRF | HEVFNATTF | 164 | 77 |
R346T
Reference QPTESIVRFPNITNLCPFGEVFNATRFASVYAWNRKRISNCVADYSVLYNS
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated QPTESIVRFPNITNLCPFHEVFNATTFASVYAWNRKRISNCVADYSVIYNF
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-B*57:01 | TRFASVYAW | TTFASVYAW | 4769 | 8 |
| HLA-B*58:01 | TRFASVYAW | TTFASVYAW | 1649 | 4 |
| HLA-B*27:05 | TRFASVYAWNRK | TTFASVYAWNRK | 127 | 22943 |
| HLA-B*38:01 | FGEVFNATRF | FHEVFNATTF | 16707 | 108 |
| HLA-B*57:01 | TRFASVYAWN | TTFASVYAWN | 21401 | 141 |
| HLA-B*58:01 | TRFASVYAWN | TTFASVYAWN | 13964 | 99 |
| HLA-A*32:01 | TRFASVYAW | TTFASVYAW | 1237 | 9 |
| HLA-A*68:01 | TRFASVYAWNRK | TTFASVYAWNRK | 1585 | 15 |
| HLA-A*11:01 | TRFASVYAWNRK | TTFASVYAWNRK | 2010 | 20 |
| HLA-B*27:05 | TRFASVYAWNR | TTFASVYAWNR | 253 | 25190 |
| HLA-A*68:01 | FGEVFNATR | FHEVFNATT | 338 | 32767 |
| HLA-A*30:01 | NATRFASVYA | NATTFASVYA | 40 | 3756 |
| HLA-A*68:02 | TRFASVYAWNRKRI | TTFASVYAWNRKRI | 20921 | 226 |
| HLA-A*30:01 | ATRFASVYA | ATTFASVYA | 3 | 275 |
| HLA-A*68:01 | TRFASVYAWNRKR | TTFASVYAWNRKR | 692 | 8 |
| HLA-B*27:05 | ATRFASVYAW | ATTFASVYAW | 325 | 24180 |
| HLA-A*68:01 | CPFGEVFNATR | CPFHEVFNATT | 457 | 32129 |
| HLA-A*30:01 | ATRFASVYAW | ATTFASVYAW | 100 | 6909 |
| HLA-A*30:01 | ATRFASVYAWNRK | ATTFASVYAWNRK | 45 | 3053 |
| HLA-B*27:05 | TRFASVYAW | TTFASVYAW | 301 | 19581 |
| HLA-A*68:01 | TRFASVYAWNR | TTFASVYAWNR | 513 | 8 |
| HLA-A*11:01 | TRFASVYAWNR | TTFASVYAWNR | 8088 | 141 |
| HLA-B*08:01 | FNATRFASV | FNATTFASV | 38 | 2163 |
| HLA-B*53:01 | TRFASVYAW | TTFASVYAW | 3827 | 71 |
| HLA-B*58:01 | VFNATRFASVYAW | VFNATTFASVYAW | 2154 | 44 |
| HLA-A*30:01 | ATRFASVY | ATTFASVY | 303 | 14033 |
| HLA-B*58:02 | TRFASVYAW | TTFASVYAW | 13911 | 307 |
| HLA-B*57:01 | VFNATRFASVYAW | VFNATTFASVYAW | 1811 | 40 |
| HLA-B*18:01 | GEVFNATRF | HEVFNATTF | 971 | 24 |
| HLA-A*30:01 | VFNATRFASVYA | VFNATTFASVYA | 112 | 4258 |
| HLA-A*03:02 | TRFASVYAWNRK | TTFASVYAWNRK | 5014 | 134 |
| HLA-A*30:01 | FNATRFASVYA | FNATTFASVYA | 316 | 10821 |
| HLA-A*03:01 | TRFASVYAWNRK | TTFASVYAWNRK | 6885 | 252 |
| HLA-A*68:02 | GEVFNATRFA | HEVFNATTFA | 879 | 50 |
| HLA-B*15:03 | GEVFNATRF | HEVFNATTF | 359 | 26 |
| HLA-C*03:02 | TRFASVYAW | TTFASVYAW | 6240 | 464 |
| HLA-B*15:03 | FGEVFNATRF | FHEVFNATTF | 694 | 60 |
| HLA-A*33:03 | TRFASVYAWNR | TTFASVYAWNR | 494 | 43 |
| HLA-C*12:03 | TRFASVYAW | TTFASVYAW | 4688 | 426 |
| HLA-B*58:01 | FNATRFASVYAW | FNATTFASVYAW | 462 | 42 |
| HLA-A*30:01 | RFASVYAWNRK | TFASVYAWNRK | 336 | 3487 |
| HLA-A*31:01 | RFASVYAWNRK | TFASVYAWNRK | 103 | 884 |
| HLA-A*68:01 | ATRFASVYAWNR | ATTFASVYAWNR | 280 | 33 |
| HLA-B*18:01 | GEVFNATRFASVY | HEVFNATTFASVY | 2165 | 265 |
| HLA-B*57:01 | FNATRFASVYAW | FNATTFASVYAW | 489 | 60 |
| HLA-A*31:01 | ATRFASVYAWNR | ATTFASVYAWNR | 19 | 152 |
| HLA-A*31:01 | ATRFASVYAWNRK | ATTFASVYAWNRK | 469 | 3463 |
| HLA-A*33:03 | TRFASVYAWNRKR | TTFASVYAWNRKR | 417 | 57 |
| HLA-A*68:01 | ATRFASVYAWNRK | ATTFASVYAWNRK | 1400 | 193 |
| HLA-A*68:01 | ATRFASVYAWNRKR | ATTFASVYAWNRKR | 1024 | 145 |
| HLA-A*02:02 | FGEVFNATRFA | FHEVFNATTFA | 2678 | 413 |
| HLA-A*31:01 | RFASVYAWNRKR | TFASVYAWNRKR | 14 | 90 |
| HLA-B*18:01 | FGEVFNATRF | FHEVFNATTF | 1074 | 179 |
| HLA-A*68:02 | ATRFASVYA | ATTFASVYA | 485 | 90 |
| HLA-A*68:01 | RFASVYAWNRKR | TFASVYAWNRKR | 1357 | 254 |
| HLA-A*68:02 | FNATRFASVYA | FNATTFASVYA | 1312 | 254 |
| HLA-A*02:06 | FGEVFNATRFA | FHEVFNATTFA | 1063 | 209 |
| HLA-A*31:01 | ATRFASVYAWNRKR | ATTFASVYAWNRKR | 78 | 363 |
| HLA-A*33:03 | RFASVYAWNRKR | TFASVYAWNRKR | 200 | 43 |
| HLA-A*68:01 | RFASVYAWNR | TFASVYAWNR | 147 | 32 |
| HLA-B*40:01 | FGEVFNATRF | FHEVFNATTF | 1994 | 435 |
| HLA-A*31:01 | NATRFASVYAWNR | NATTFASVYAWNR | 263 | 1165 |
| HLA-A*03:01 | ATRFASVYAWNRK | ATTFASVYAWNRK | 151 | 656 |
| HLA-B*40:02 | FGEVFNATRF | FHEVFNATTF | 1615 | 380 |
| HLA-A*31:01 | RFASVYAWNR | TFASVYAWNR | 7 | 29 |
| HLA-A*23:01 | TRFASVYAW | TTFASVYAW | 1649 | 403 |
| HLA-A*68:02 | FGEVFNATRFA | FHEVFNATTFA | 939 | 230 |
| HLA-B*57:01 | RFASVYAW | TFASVYAW | 586 | 144 |
| HLA-A*33:03 | RFASVYAWNRK | TFASVYAWNRK | 1936 | 492 |
| HLA-B*35:01 | CPFGEVFNATRF | CPFHEVFNATTF | 1090 | 291 |
| HLA-A*68:02 | NATRFASVYA | NATTFASVYA | 204 | 55 |
| HLA-B*58:01 | RFASVYAW | TFASVYAW | 296 | 83 |
| HLA-A*24:02 | RFASVYAW | TFASVYAW | 446 | 1553 |
| HLA-A*33:03 | RFASVYAWNR | TFASVYAWNR | 38 | 11 |
| HLA-A*23:01 | RFASVYAW | TFASVYAW | 277 | 951 |
| HLA-A*68:01 | NATRFASVYAWNR | NATTFASVYAWNR | 444 | 130 |
| HLA-A*11:01 | ATRFASVYAWNRK | ATTFASVYAWNRK | 130 | 39 |
| HLA-A*68:01 | EVFNATRFA | EVFNATTFA | 579 | 181 |
| HLA-B*40:02 | GEVFNATRF | HEVFNATTF | 283 | 96 |
| HLA-B*35:01 | FNATRFASVY | FNATTFASVY | 106 | 36 |
| HLA-A*68:02 | EVFNATRFA | EVFNATTFA | 14 | 5 |
| HLA-A*11:01 | ATRFASVYAWNR | ATTFASVYAWNR | 733 | 271 |
| HLA-A*68:01 | EVFNATRFASVY | EVFNATTFASVY | 407 | 155 |
| HLA-A*68:02 | FNATRFASV | FNATTFASV | 96 | 38 |
| HLA-A*29:02 | EVFNATRFASVY | EVFNATTFASVY | 1045 | 415 |
| HLA-B*15:03 | GEVFNATRFASVY | HEVFNATTFASVY | 1058 | 423 |
| HLA-A*23:01 | PFGEVFNATRF | PFHEVFNATTF | 1002 | 401 |
| HLA-A*29:02 | VFNATRFASVY | VFNATTFASVY | 130 | 53 |
| HLA-B*15:03 | TRFASVYAW | TTFASVYAW | 224 | 496 |
| HLA-A*31:01 | TRFASVYAWNRKR | TTFASVYAWNRKR | 213 | 98 |
| HLA-A*02:06 | FNATRFASV | FNATTFASV | 210 | 98 |
| HLA-B*40:01 | GEVFNATRF | HEVFNATTF | 164 | 77 |
| HLA-A*68:01 | FNATRFASVYAWNR | FNATTFASVYAWNR | 934 | 442 |
| HLA-A*03:02 | ATRFASVYAWNRK | ATTFASVYAWNRK | 493 | 242 |
| HLA-A*02:02 | FNATRFASV | FNATTFASV | 408 | 202 |
L368I
Reference NATRFASVYAWNRKRISNCVADYSVLYNSASFSTFKCYGVSPTKLNDLCFT
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated NATTFASVYAWNRKRISNCVADYSVIYNFAPFFAFKCYGVSPTKLNDLCFT
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-A*32:01 | VLYNSASFS | VIYNFAPFF | 22186 | 48 |
| HLA-A*23:01 | LYNSASFS | IYNFAPFF | 22508 | 99 |
| HLA-A*24:02 | LYNSASFS | IYNFAPFF | 25164 | 136 |
| HLA-A*23:01 | VLYNSASFS | VIYNFAPFF | 32767 | 178 |
| HLA-A*29:02 | VLYNSASFS | VIYNFAPFF | 10439 | 76 |
| HLA-C*03:02 | VLYNSASFS | VIYNFAPFF | 13004 | 100 |
| HLA-B*15:03 | VLYNSASFS | VIYNFAPFF | 3280 | 27 |
| HLA-A*26:01 | SVLYNSASFS | SVIYNFAPFF | 21993 | 202 |
| HLA-C*14:02 | LYNSASFS | IYNFAPFF | 5693 | 80 |
| HLA-C*05:01 | VADYSVLYNS | VADYSVIYNF | 11802 | 172 |
| HLA-A*32:01 | SVLYNSASFS | SVIYNFAPFF | 14340 | 227 |
| HLA-A*29:02 | SVLYNSASFS | SVIYNFAPFF | 16185 | 274 |
| HLA-B*15:01 | SVLYNSASFS | SVIYNFAPFF | 2838 | 50 |
| HLA-A*26:01 | SVLYNSASF | SVIYNFAPF | 1313 | 24 |
| HLA-A*23:01 | SVLYNSASFS | SVIYNFAPFF | 24524 | 498 |
| HLA-C*03:02 | SVLYNSASFS | SVIYNFAPFF | 8888 | 192 |
| HLA-A*68:02 | SVLYNSASFST | SVIYNFAPFFA | 14147 | 319 |
| HLA-B*58:01 | VADYSVLYNS | VADYSVIYNF | 6124 | 141 |
| HLA-C*16:01 | VLYNSASFS | VIYNFAPFF | 18639 | 432 |
| HLA-B*15:03 | SVLYNSASFS | SVIYNFAPFF | 2850 | 68 |
| HLA-A*23:01 | DYSVLYNSASFS | DYSVIYNFAPFF | 14260 | 341 |
| HLA-C*03:02 | VADYSVLYNS | VADYSVIYNF | 20606 | 498 |
| HLA-A*68:02 | VLYNSASFST | VIYNFAPFFA | 11417 | 297 |
| HLA-A*25:01 | SVLYNSASF | SVIYNFAPF | 2252 | 59 |
| HLA-C*12:03 | VLYNSASFS | VIYNFAPFF | 12383 | 333 |
| HLA-C*14:02 | VLYNSASFS | VIYNFAPFF | 11085 | 360 |
| HLA-A*68:02 | SVLYNSASF | SVIYNFAPF | 6927 | 241 |
| HLA-B*15:01 | VLYNSASFS | VIYNFAPFF | 8189 | 317 |
| HLA-B*57:01 | VADYSVLYNS | VADYSVIYNF | 11273 | 446 |
| HLA-B*58:01 | YSVLYNSASFS | YSVIYNFAPFF | 6920 | 277 |
| HLA-A*02:06 | SVLYNSASF | SVIYNFAPF | 2112 | 98 |
| HLA-A*02:06 | SVLYNSASFST | SVIYNFAPFFA | 4097 | 193 |
| HLA-A*68:01 | SVLYNSASF | SVIYNFAPF | 6337 | 311 |
| HLA-B*15:01 | VLYNSASFSTF | VIYNFAPFFAF | 50 | 951 |
| HLA-B*40:02 | ADYSVLYNS | ADYSVIYNF | 4645 | 272 |
| HLA-B*15:03 | YSVLYNSASFS | YSVIYNFAPFF | 3170 | 196 |
| HLA-A*11:01 | LYNSASFSTFK | IYNFAPFFAFK | 43 | 681 |
| HLA-A*26:01 | YSVLYNSASF | YSVIYNFAPF | 1544 | 107 |
| HLA-A*02:06 | VLYNSASFST | VIYNFAPFFA | 710 | 58 |
| HLA-A*30:01 | LYNSASFST | IYNFAPFFA | 3467 | 316 |
| HLA-C*03:02 | YSVLYNSASFS | YSVIYNFAPFF | 2405 | 260 |
| HLA-A*02:11 | SVLYNSASFST | SVIYNFAPFFA | 3830 | 425 |
| HLA-A*33:03 | SVLYNSASFSTFK | SVIYNFAPFFAFK | 2778 | 320 |
| HLA-C*12:02 | SVLYNSASF | SVIYNFAPF | 507 | 63 |
| HLA-A*25:01 | YSVLYNSASF | YSVIYNFAPF | 2498 | 338 |
| HLA-A*23:01 | SVLYNSASF | SVIYNFAPF | 990 | 145 |
| HLA-A*30:02 | VLYNSASFS | VIYNFAPFF | 2919 | 448 |
| HLA-A*02:06 | YSVLYNSASF | YSVIYNFAPF | 2478 | 397 |
| HLA-B*15:03 | VLYNSASFSTF | VIYNFAPFFAF | 38 | 235 |
| HLA-A*29:02 | LYNSASFSTF | IYNFAPFFAF | 387 | 63 |
| HLA-A*29:02 | SVLYNSASF | SVIYNFAPF | 1130 | 186 |
| HLA-C*12:03 | SVLYNSASF | SVIYNFAPF | 748 | 127 |
| HLA-A*29:02 | SNCVADYSVLY | SNCVADYSVIY | 83 | 483 |
| HLA-A*31:01 | SVLYNSASFSTFK | SVIYNFAPFFAFK | 857 | 150 |
| HLA-A*02:01 | VLYNSASFST | VIYNFAPFFA | 243 | 43 |
| HLA-A*29:02 | NCVADYSVLY | NCVADYSVIY | 57 | 320 |
| HLA-A*29:02 | ISNCVADYSVLY | ISNCVADYSVIY | 116 | 625 |
| HLA-A*03:01 | VLYNSASFSTFKC | VIYNFAPFFAFKC | 312 | 1619 |
| HLA-A*29:02 | RISNCVADYSVLY | RISNCVADYSVIY | 249 | 1271 |
| HLA-C*02:02 | SVLYNSASF | SVIYNFAPF | 1090 | 218 |
| HLA-C*02:10 | SVLYNSASF | SVIYNFAPF | 1090 | 218 |
| HLA-A*01:01 | CVADYSVLY | CVADYSVIY | 76 | 367 |
| HLA-A*24:02 | SVLYNSASF | SVIYNFAPF | 1977 | 410 |
| HLA-A*03:01 | LYNSASFSTFK | IYNFAPFFAFK | 295 | 1419 |
| HLA-A*01:01 | VADYSVLY | VADYSVIY | 339 | 1549 |
| HLA-B*15:01 | SVLYNSASF | SVIYNFAPF | 52 | 12 |
| HLA-B*35:01 | SVLYNSASF | SVIYNFAPF | 153 | 36 |
| HLA-A*02:02 | VLYNSASFST | VIYNFAPFFA | 202 | 49 |
| HLA-A*31:01 | VLYNSASFSTFK | VIYNFAPFFAFK | 565 | 139 |
| HLA-A*11:01 | CVADYSVLY | CVADYSVIY | 169 | 673 |
| HLA-A*68:01 | CVADYSVLY | CVADYSVIY | 78 | 304 |
| HLA-A*03:01 | VLYNSASFSTFK | VIYNFAPFFAFK | 7 | 27 |
| HLA-A*02:11 | VLYNSASFST | VIYNFAPFFA | 77 | 20 |
| HLA-A*68:01 | NCVADYSVLY | NCVADYSVIY | 267 | 1017 |
| HLA-A*03:02 | LYNSASFSTFK | IYNFAPFFAFK | 202 | 760 |
| HLA-A*68:02 | CVADYSVLYNSA | CVADYSVIYNFA | 1306 | 349 |
| HLA-A*31:01 | LYNSASFSTFK | IYNFAPFFAFK | 299 | 81 |
| HLA-B*15:01 | YSVLYNSASFSTF | YSVIYNFAPFFAF | 164 | 604 |
| HLA-A*29:02 | YSVLYNSASF | YSVIYNFAPF | 1696 | 462 |
| HLA-A*30:02 | VLYNSASFSTFKCY | VIYNFAPFFAFKCY | 272 | 973 |
| HLA-B*15:01 | LYNSASFSTF | IYNFAPFFAF | 468 | 1565 |
| HLA-C*03:02 | SVLYNSASF | SVIYNFAPF | 29 | 9 |
| HLA-A*23:01 | YSVLYNSASF | YSVIYNFAPF | 1120 | 364 |
| HLA-B*27:05 | KRISNCVADYSVLY | KRISNCVADYSVIY | 358 | 1096 |
| HLA-A*26:01 | NCVADYSVLY | NCVADYSVIY | 183 | 556 |
| HLA-A*29:02 | CVADYSVLY | CVADYSVIY | 6 | 18 |
| HLA-C*03:02 | YSVLYNSASFSTF | YSVIYNFAPFFAF | 113 | 331 |
| HLA-A*26:01 | CVADYSVLY | CVADYSVIY | 33 | 95 |
| HLA-A*29:02 | LYNSASFSTFKCY | IYNFAPFFAFKCY | 297 | 105 |
| HLA-A*68:01 | VLYNSASFSTFK | VIYNFAPFFAFK | 257 | 91 |
| HLA-A*03:02 | VLYNSASFSTFK | VIYNFAPFFAFK | 13 | 36 |
| HLA-C*14:02 | SVLYNSASF | SVIYNFAPF | 152 | 56 |
| HLA-B*15:03 | VLYNSASF | VIYNFAPF | 34 | 92 |
| HLA-B*58:01 | YSVLYNSASFSTF | YSVIYNFAPFFAF | 429 | 1153 |
| HLA-A*01:01 | ISNCVADYSVLY | ISNCVADYSVIY | 123 | 327 |
| HLA-C*03:03 | SVLYNSASF | SVIYNFAPF | 114 | 45 |
| HLA-C*03:04 | SVLYNSASF | SVIYNFAPF | 114 | 45 |
| HLA-A*02:11 | VLYNSASFS | VIYNFAPFF | 258 | 645 |
| HLA-A*23:01 | DYSVLYNSASF | DYSVIYNFAPF | 1039 | 424 |
| HLA-A*33:03 | LYNSASFSTFK | IYNFAPFFAFK | 1158 | 476 |
| HLA-B*35:01 | YSVLYNSASF | YSVIYNFAPF | 328 | 136 |
| HLA-A*30:02 | SVLYNSASF | SVIYNFAPF | 1057 | 444 |
| HLA-A*23:01 | SVLYNSASFSTF | SVIYNFAPFFAF | 656 | 277 |
| HLA-B*15:01 | VLYNSASF | VIYNFAPF | 224 | 518 |
| HLA-A*30:02 | SNCVADYSVLY | SNCVADYSVIY | 427 | 982 |
| HLA-A*30:02 | CVADYSVLY | CVADYSVIY | 72 | 159 |
| HLA-C*14:02 | LYNSASFSTF | IYNFAPFFAF | 20 | 43 |
| HLA-A*68:01 | YSVLYNSASFSTFK | YSVIYNFAPFFAFK | 144 | 67 |
| HLA-A*30:02 | KRISNCVADYSVLY | KRISNCVADYSVIY | 456 | 969 |
| HLA-A*30:02 | RISNCVADYSVLY | RISNCVADYSVIY | 81 | 170 |
| HLA-A*68:01 | SVLYNSASFSTFK | SVIYNFAPFFAFK | 41 | 20 |
| HLA-A*03:01 | SVLYNSASFSTFK | SVIYNFAPFFAFK | 47 | 96 |
S371F
Reference RFASVYAWNRKRISNCVADYSVLYNSASFSTFKCYGVSPTKLNDLCFTNVY
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated TFASVYAWNRKRISNCVADYSVIYNFAPFFAFKCYGVSPTKLNDLCFTNVY
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-A*32:01 | VLYNSASFS | VIYNFAPFF | 22186 | 48 |
| HLA-A*23:01 | LYNSASFS | IYNFAPFF | 22508 | 99 |
| HLA-A*24:02 | LYNSASFS | IYNFAPFF | 25164 | 136 |
| HLA-A*23:01 | VLYNSASFS | VIYNFAPFF | 32767 | 178 |
| HLA-A*29:02 | VLYNSASFS | VIYNFAPFF | 10439 | 76 |
| HLA-C*03:02 | VLYNSASFS | VIYNFAPFF | 13004 | 100 |
| HLA-B*15:03 | VLYNSASFS | VIYNFAPFF | 3280 | 27 |
| HLA-A*26:01 | SVLYNSASFS | SVIYNFAPFF | 21993 | 202 |
| HLA-C*14:02 | LYNSASFS | IYNFAPFF | 5693 | 80 |
| HLA-C*05:01 | VADYSVLYNS | VADYSVIYNF | 11802 | 172 |
| HLA-A*32:01 | SVLYNSASFS | SVIYNFAPFF | 14340 | 227 |
| HLA-A*29:02 | SVLYNSASFS | SVIYNFAPFF | 16185 | 274 |
| HLA-B*15:01 | SVLYNSASFS | SVIYNFAPFF | 2838 | 50 |
| HLA-A*26:01 | SVLYNSASF | SVIYNFAPF | 1313 | 24 |
| HLA-A*23:01 | SVLYNSASFS | SVIYNFAPFF | 24524 | 498 |
| HLA-C*03:02 | SVLYNSASFS | SVIYNFAPFF | 8888 | 192 |
| HLA-A*68:02 | SVLYNSASFST | SVIYNFAPFFA | 14147 | 319 |
| HLA-B*58:01 | VADYSVLYNS | VADYSVIYNF | 6124 | 141 |
| HLA-C*16:01 | VLYNSASFS | VIYNFAPFF | 18639 | 432 |
| HLA-B*15:03 | SVLYNSASFS | SVIYNFAPFF | 2850 | 68 |
| HLA-A*23:01 | DYSVLYNSASFS | DYSVIYNFAPFF | 14260 | 341 |
| HLA-C*03:02 | VADYSVLYNS | VADYSVIYNF | 20606 | 498 |
| HLA-A*01:01 | NSASFSTFKCY | NFAPFFAFKCY | 328 | 13051 |
| HLA-A*68:02 | VLYNSASFST | VIYNFAPFFA | 11417 | 297 |
| HLA-A*25:01 | SVLYNSASF | SVIYNFAPF | 2252 | 59 |
| HLA-C*12:03 | VLYNSASFS | VIYNFAPFF | 12383 | 333 |
| HLA-C*16:01 | NSASFSTF | NFAPFFAF | 243 | 8562 |
| HLA-C*14:02 | VLYNSASFS | VIYNFAPFF | 11085 | 360 |
| HLA-A*23:01 | NSASFSTF | NFAPFFAF | 12240 | 424 |
| HLA-A*68:02 | SVLYNSASF | SVIYNFAPF | 6927 | 241 |
| HLA-A*29:02 | YNSASFSTF | YNFAPFFAF | 1671 | 60 |
| HLA-B*15:01 | VLYNSASFS | VIYNFAPFF | 8189 | 317 |
| HLA-B*57:01 | VADYSVLYNS | VADYSVIYNF | 11273 | 446 |
| HLA-B*58:01 | YSVLYNSASFS | YSVIYNFAPFF | 6920 | 277 |
| HLA-A*68:02 | NSASFSTFKCYGV | NFAPFFAFKCYGV | 48 | 1043 |
| HLA-A*02:06 | SVLYNSASF | SVIYNFAPF | 2112 | 98 |
| HLA-A*02:06 | SVLYNSASFST | SVIYNFAPFFA | 4097 | 193 |
| HLA-C*14:02 | NSASFSTF | NFAPFFAF | 2544 | 120 |
| HLA-A*68:01 | SVLYNSASF | SVIYNFAPF | 6337 | 311 |
| HLA-B*15:01 | VLYNSASFSTF | VIYNFAPFFAF | 50 | 951 |
| HLA-A*30:02 | SASFSTFKCY | FAPFFAFKCY | 71 | 1241 |
| HLA-B*40:02 | ADYSVLYNS | ADYSVIYNF | 4645 | 272 |
| HLA-B*15:03 | YSVLYNSASFS | YSVIYNFAPFF | 3170 | 196 |
| HLA-A*11:01 | LYNSASFSTFK | IYNFAPFFAFK | 43 | 681 |
| HLA-A*32:01 | YNSASFSTF | YNFAPFFAF | 4333 | 294 |
| HLA-A*33:03 | YNSASFSTFK | YNFAPFFAFK | 823 | 57 |
| HLA-A*26:01 | YSVLYNSASF | YSVIYNFAPF | 1544 | 107 |
| HLA-B*18:01 | YNSASFSTF | YNFAPFFAF | 3495 | 284 |
| HLA-A*02:06 | VLYNSASFST | VIYNFAPFFA | 710 | 58 |
| HLA-A*33:03 | NSASFSTFK | NFAPFFAFK | 181 | 16 |
| HLA-A*30:01 | LYNSASFST | IYNFAPFFA | 3467 | 316 |
| HLA-A*11:01 | NSASFSTFK | NFAPFFAFK | 9 | 94 |
| HLA-A*29:02 | NSASFSTFKCY | NFAPFFAFKCY | 662 | 64 |
| HLA-A*29:02 | YNSASFSTFKCY | YNFAPFFAFKCY | 842 | 86 |
| HLA-C*03:02 | YSVLYNSASFS | YSVIYNFAPFF | 2405 | 260 |
| HLA-A*02:11 | SVLYNSASFST | SVIYNFAPFFA | 3830 | 425 |
| HLA-A*33:03 | SVLYNSASFSTFK | SVIYNFAPFFAFK | 2778 | 320 |
| HLA-A*31:01 | YNSASFSTFK | YNFAPFFAFK | 607 | 73 |
| HLA-C*12:02 | SVLYNSASF | SVIYNFAPF | 507 | 63 |
| HLA-B*35:01 | SASFSTFKCY | FAPFFAFKCY | 490 | 65 |
| HLA-A*25:01 | YSVLYNSASF | YSVIYNFAPF | 2498 | 338 |
| HLA-A*23:01 | SVLYNSASF | SVIYNFAPF | 990 | 145 |
| HLA-A*30:02 | VLYNSASFS | VIYNFAPFF | 2919 | 448 |
| HLA-A*02:06 | YSVLYNSASF | YSVIYNFAPF | 2478 | 397 |
| HLA-A*31:01 | NSASFSTFK | NFAPFFAFK | 293 | 47 |
| HLA-B*15:03 | VLYNSASFSTF | VIYNFAPFFAF | 38 | 235 |
| HLA-A*29:02 | LYNSASFSTF | IYNFAPFFAF | 387 | 63 |
| HLA-A*29:02 | SVLYNSASF | SVIYNFAPF | 1130 | 186 |
| HLA-C*12:03 | SVLYNSASF | SVIYNFAPF | 748 | 127 |
| HLA-A*31:01 | SVLYNSASFSTFK | SVIYNFAPFFAFK | 857 | 150 |
| HLA-A*02:01 | VLYNSASFST | VIYNFAPFFA | 243 | 43 |
| HLA-A*23:01 | YNSASFSTF | YNFAPFFAF | 352 | 63 |
| HLA-A*03:01 | VLYNSASFSTFKC | VIYNFAPFFAFKC | 312 | 1619 |
| HLA-C*02:02 | SVLYNSASF | SVIYNFAPF | 1090 | 218 |
| HLA-C*02:10 | SVLYNSASF | SVIYNFAPF | 1090 | 218 |
| HLA-A*24:02 | SVLYNSASF | SVIYNFAPF | 1977 | 410 |
| HLA-A*03:01 | LYNSASFSTFK | IYNFAPFFAFK | 295 | 1419 |
| HLA-A*03:01 | NSASFSTFK | NFAPFFAFK | 113 | 494 |
| HLA-B*15:01 | SVLYNSASF | SVIYNFAPF | 52 | 12 |
| HLA-B*35:01 | SVLYNSASF | SVIYNFAPF | 153 | 36 |
| HLA-B*35:01 | YNSASFSTF | YNFAPFFAF | 760 | 180 |
| HLA-A*02:02 | VLYNSASFST | VIYNFAPFFA | 202 | 49 |
| HLA-A*31:01 | VLYNSASFSTFK | VIYNFAPFFAFK | 565 | 139 |
| HLA-A*01:01 | SASFSTFKCY | FAPFFAFKCY | 461 | 1849 |
| HLA-A*11:01 | SASFSTFK | FAPFFAFK | 452 | 1795 |
| HLA-A*03:01 | VLYNSASFSTFK | VIYNFAPFFAFK | 7 | 27 |
| HLA-A*02:11 | VLYNSASFST | VIYNFAPFFA | 77 | 20 |
| HLA-A*68:01 | SASFSTFK | FAPFFAFK | 1114 | 290 |
| HLA-A*03:02 | LYNSASFSTFK | IYNFAPFFAFK | 202 | 760 |
| HLA-A*68:02 | CVADYSVLYNSA | CVADYSVIYNFA | 1306 | 349 |
| HLA-A*31:01 | LYNSASFSTFK | IYNFAPFFAFK | 299 | 81 |
| HLA-B*15:01 | YSVLYNSASFSTF | YSVIYNFAPFFAF | 164 | 604 |
| HLA-A*29:02 | YSVLYNSASF | YSVIYNFAPF | 1696 | 462 |
| HLA-A*03:02 | NSASFSTFK | NFAPFFAFK | 37 | 135 |
| HLA-A*30:02 | NSASFSTFKCY | NFAPFFAFKCY | 387 | 1395 |
| HLA-C*12:03 | SASFSTFKCY | FAPFFAFKCY | 498 | 139 |
| HLA-A*30:02 | VLYNSASFSTFKCY | VIYNFAPFFAFKCY | 272 | 973 |
| HLA-B*15:01 | LYNSASFSTF | IYNFAPFFAF | 468 | 1565 |
| HLA-A*68:01 | NSASFSTFK | NFAPFFAFK | 4 | 13 |
| HLA-C*03:02 | SVLYNSASF | SVIYNFAPF | 29 | 9 |
| HLA-A*11:01 | YNSASFSTFK | YNFAPFFAFK | 12 | 38 |
| HLA-A*23:01 | YSVLYNSASF | YSVIYNFAPF | 1120 | 364 |
| HLA-A*02:06 | SASFSTFKCYGV | FAPFFAFKCYGV | 1416 | 481 |
| HLA-C*03:02 | YSVLYNSASFSTF | YSVIYNFAPFFAF | 113 | 331 |
| HLA-A*29:02 | LYNSASFSTFKCY | IYNFAPFFAFKCY | 297 | 105 |
| HLA-A*68:01 | VLYNSASFSTFK | VIYNFAPFFAFK | 257 | 91 |
| HLA-A*03:02 | VLYNSASFSTFK | VIYNFAPFFAFK | 13 | 36 |
| HLA-C*14:02 | SVLYNSASF | SVIYNFAPF | 152 | 56 |
| HLA-B*15:03 | VLYNSASF | VIYNFAPF | 34 | 92 |
| HLA-B*58:01 | YSVLYNSASFSTF | YSVIYNFAPFFAF | 429 | 1153 |
| HLA-C*12:02 | SASFSTFKCY | FAPFFAFKCY | 710 | 275 |
| HLA-C*03:03 | SVLYNSASF | SVIYNFAPF | 114 | 45 |
| HLA-C*03:04 | SVLYNSASF | SVIYNFAPF | 114 | 45 |
| HLA-A*02:11 | VLYNSASFS | VIYNFAPFF | 258 | 645 |
| HLA-A*68:01 | NSASFSTFKC | NFAPFFAFKC | 281 | 702 |
| HLA-A*23:01 | DYSVLYNSASF | DYSVIYNFAPF | 1039 | 424 |
| HLA-A*33:03 | LYNSASFSTFK | IYNFAPFFAFK | 1158 | 476 |
| HLA-A*03:01 | YNSASFSTFK | YNFAPFFAFK | 80 | 194 |
| HLA-B*35:01 | YSVLYNSASF | YSVIYNFAPF | 328 | 136 |
| HLA-A*30:02 | SVLYNSASF | SVIYNFAPF | 1057 | 444 |
| HLA-A*23:01 | SVLYNSASFSTF | SVIYNFAPFFAF | 656 | 277 |
| HLA-B*15:01 | VLYNSASF | VIYNFAPF | 224 | 518 |
| HLA-B*15:01 | SASFSTFKCY | FAPFFAFKCY | 256 | 561 |
| HLA-C*03:02 | SASFSTFKCY | FAPFFAFKCY | 212 | 97 |
| HLA-C*14:02 | LYNSASFSTF | IYNFAPFFAF | 20 | 43 |
| HLA-A*68:01 | YSVLYNSASFSTFK | YSVIYNFAPFFAFK | 144 | 67 |
| HLA-A*68:01 | SVLYNSASFSTFK | SVIYNFAPFFAFK | 41 | 20 |
| HLA-A*03:01 | SVLYNSASFSTFK | SVIYNFAPFFAFK | 47 | 96 |
S373P
Reference ASVYAWNRKRISNCVADYSVLYNSASFSTFKCYGVSPTKLNDLCFTNVYAD
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated ASVYAWNRKRISNCVADYSVIYNFAPFFAFKCYGVSPTKLNDLCFTNVYAD
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-A*32:01 | VLYNSASFS | VIYNFAPFF | 22186 | 48 |
| HLA-A*23:01 | LYNSASFS | IYNFAPFF | 22508 | 99 |
| HLA-A*24:02 | LYNSASFS | IYNFAPFF | 25164 | 136 |
| HLA-A*23:01 | VLYNSASFS | VIYNFAPFF | 32767 | 178 |
| HLA-A*29:02 | VLYNSASFS | VIYNFAPFF | 10439 | 76 |
| HLA-C*03:02 | VLYNSASFS | VIYNFAPFF | 13004 | 100 |
| HLA-B*15:03 | VLYNSASFS | VIYNFAPFF | 3280 | 27 |
| HLA-A*26:01 | SVLYNSASFS | SVIYNFAPFF | 21993 | 202 |
| HLA-C*14:02 | LYNSASFS | IYNFAPFF | 5693 | 80 |
| HLA-A*32:01 | SVLYNSASFS | SVIYNFAPFF | 14340 | 227 |
| HLA-A*30:02 | ASFSTFKCY | APFFAFKCY | 43 | 2647 |
| HLA-A*29:02 | SVLYNSASFS | SVIYNFAPFF | 16185 | 274 |
| HLA-B*15:01 | ASFSTFKCY | APFFAFKCY | 107 | 6085 |
| HLA-B*15:01 | SVLYNSASFS | SVIYNFAPFF | 2838 | 50 |
| HLA-A*26:01 | SVLYNSASF | SVIYNFAPF | 1313 | 24 |
| HLA-A*02:06 | ASFSTFKCYGV | APFFAFKCYGV | 231 | 11537 |
| HLA-A*23:01 | SVLYNSASFS | SVIYNFAPFF | 24524 | 498 |
| HLA-C*03:02 | SVLYNSASFS | SVIYNFAPFF | 8888 | 192 |
| HLA-C*16:01 | ASFSTFKCY | APFFAFKCY | 68 | 3117 |
| HLA-A*68:02 | SVLYNSASFST | SVIYNFAPFFA | 14147 | 319 |
| HLA-C*12:03 | ASFSTFKCY | APFFAFKCY | 115 | 5066 |
| HLA-C*16:01 | VLYNSASFS | VIYNFAPFF | 18639 | 432 |
| HLA-B*15:03 | SVLYNSASFS | SVIYNFAPFF | 2850 | 68 |
| HLA-A*23:01 | DYSVLYNSASFS | DYSVIYNFAPFF | 14260 | 341 |
| HLA-C*03:02 | ASFSTFKCY | APFFAFKCY | 64 | 2656 |
| HLA-A*01:01 | NSASFSTFKCY | NFAPFFAFKCY | 328 | 13051 |
| HLA-A*68:02 | VLYNSASFST | VIYNFAPFFA | 11417 | 297 |
| HLA-A*25:01 | SVLYNSASF | SVIYNFAPF | 2252 | 59 |
| HLA-C*12:03 | VLYNSASFS | VIYNFAPFF | 12383 | 333 |
| HLA-C*16:01 | NSASFSTF | NFAPFFAF | 243 | 8562 |
| HLA-A*11:01 | ASFSTFKCY | APFFAFKCY | 379 | 12606 |
| HLA-C*14:02 | VLYNSASFS | VIYNFAPFF | 11085 | 360 |
| HLA-A*23:01 | NSASFSTF | NFAPFFAF | 12240 | 424 |
| HLA-A*68:02 | SVLYNSASF | SVIYNFAPF | 6927 | 241 |
| HLA-A*29:02 | YNSASFSTF | YNFAPFFAF | 1671 | 60 |
| HLA-B*15:01 | VLYNSASFS | VIYNFAPFF | 8189 | 317 |
| HLA-B*35:01 | ASFSTFKCY | APFFAFKCY | 2345 | 92 |
| HLA-B*58:01 | YSVLYNSASFS | YSVIYNFAPFF | 6920 | 277 |
| HLA-C*12:02 | ASFSTFKCY | APFFAFKCY | 338 | 8222 |
| HLA-A*68:02 | NSASFSTFKCYGV | NFAPFFAFKCYGV | 48 | 1043 |
| HLA-A*02:06 | SVLYNSASF | SVIYNFAPF | 2112 | 98 |
| HLA-A*02:06 | SVLYNSASFST | SVIYNFAPFFA | 4097 | 193 |
| HLA-C*14:02 | NSASFSTF | NFAPFFAF | 2544 | 120 |
| HLA-B*15:03 | ASFSTFKCY | APFFAFKCY | 25 | 529 |
| HLA-A*68:01 | SVLYNSASF | SVIYNFAPF | 6337 | 311 |
| HLA-B*15:01 | VLYNSASFSTF | VIYNFAPFFAF | 50 | 951 |
| HLA-A*30:02 | SASFSTFKCY | FAPFFAFKCY | 71 | 1241 |
| HLA-C*14:02 | SFSTFKCY | PFFAFKCY | 386 | 6449 |
| HLA-B*15:03 | YSVLYNSASFS | YSVIYNFAPFF | 3170 | 196 |
| HLA-A*11:01 | LYNSASFSTFK | IYNFAPFFAFK | 43 | 681 |
| HLA-A*32:01 | YNSASFSTF | YNFAPFFAF | 4333 | 294 |
| HLA-A*33:03 | YNSASFSTFK | YNFAPFFAFK | 823 | 57 |
| HLA-A*26:01 | YSVLYNSASF | YSVIYNFAPF | 1544 | 107 |
| HLA-B*18:01 | YNSASFSTF | YNFAPFFAF | 3495 | 284 |
| HLA-A*02:06 | VLYNSASFST | VIYNFAPFFA | 710 | 58 |
| HLA-A*33:03 | NSASFSTFK | NFAPFFAFK | 181 | 16 |
| HLA-A*30:01 | LYNSASFST | IYNFAPFFA | 3467 | 316 |
| HLA-A*11:01 | NSASFSTFK | NFAPFFAFK | 9 | 94 |
| HLA-A*29:02 | NSASFSTFKCY | NFAPFFAFKCY | 662 | 64 |
| HLA-A*29:02 | YNSASFSTFKCY | YNFAPFFAFKCY | 842 | 86 |
| HLA-C*03:02 | YSVLYNSASFS | YSVIYNFAPFF | 2405 | 260 |
| HLA-A*29:02 | ASFSTFKCY | APFFAFKCY | 201 | 1830 |
| HLA-A*02:11 | SVLYNSASFST | SVIYNFAPFFA | 3830 | 425 |
| HLA-A*33:03 | SVLYNSASFSTFK | SVIYNFAPFFAFK | 2778 | 320 |
| HLA-A*31:01 | YNSASFSTFK | YNFAPFFAFK | 607 | 73 |
| HLA-C*12:02 | SVLYNSASF | SVIYNFAPF | 507 | 63 |
| HLA-B*35:01 | SASFSTFKCY | FAPFFAFKCY | 490 | 65 |
| HLA-A*25:01 | YSVLYNSASF | YSVIYNFAPF | 2498 | 338 |
| HLA-A*23:01 | SVLYNSASF | SVIYNFAPF | 990 | 145 |
| HLA-A*30:02 | VLYNSASFS | VIYNFAPFF | 2919 | 448 |
| HLA-A*02:06 | YSVLYNSASF | YSVIYNFAPF | 2478 | 397 |
| HLA-A*31:01 | NSASFSTFK | NFAPFFAFK | 293 | 47 |
| HLA-B*15:03 | VLYNSASFSTF | VIYNFAPFFAF | 38 | 235 |
| HLA-A*29:02 | LYNSASFSTF | IYNFAPFFAF | 387 | 63 |
| HLA-A*29:02 | SVLYNSASF | SVIYNFAPF | 1130 | 186 |
| HLA-C*12:03 | SVLYNSASF | SVIYNFAPF | 748 | 127 |
| HLA-A*31:01 | SVLYNSASFSTFK | SVIYNFAPFFAFK | 857 | 150 |
| HLA-A*02:01 | VLYNSASFST | VIYNFAPFFA | 243 | 43 |
| HLA-A*23:01 | YNSASFSTF | YNFAPFFAF | 352 | 63 |
| HLA-A*03:01 | VLYNSASFSTFKC | VIYNFAPFFAFKC | 312 | 1619 |
| HLA-C*02:02 | SVLYNSASF | SVIYNFAPF | 1090 | 218 |
| HLA-C*02:10 | SVLYNSASF | SVIYNFAPF | 1090 | 218 |
| HLA-A*24:02 | SVLYNSASF | SVIYNFAPF | 1977 | 410 |
| HLA-A*03:01 | LYNSASFSTFK | IYNFAPFFAFK | 295 | 1419 |
| HLA-A*03:01 | NSASFSTFK | NFAPFFAFK | 113 | 494 |
| HLA-B*15:01 | SVLYNSASF | SVIYNFAPF | 52 | 12 |
| HLA-B*35:01 | SVLYNSASF | SVIYNFAPF | 153 | 36 |
| HLA-B*35:01 | YNSASFSTF | YNFAPFFAF | 760 | 180 |
| HLA-A*02:02 | VLYNSASFST | VIYNFAPFFA | 202 | 49 |
| HLA-A*31:01 | VLYNSASFSTFK | VIYNFAPFFAFK | 565 | 139 |
| HLA-A*01:01 | SASFSTFKCY | FAPFFAFKCY | 461 | 1849 |
| HLA-A*11:01 | SASFSTFK | FAPFFAFK | 452 | 1795 |
| HLA-A*03:01 | VLYNSASFSTFK | VIYNFAPFFAFK | 7 | 27 |
| HLA-A*02:11 | VLYNSASFST | VIYNFAPFFA | 77 | 20 |
| HLA-A*68:01 | SASFSTFK | FAPFFAFK | 1114 | 290 |
| HLA-A*03:02 | LYNSASFSTFK | IYNFAPFFAFK | 202 | 760 |
| HLA-A*31:01 | LYNSASFSTFK | IYNFAPFFAFK | 299 | 81 |
| HLA-B*15:01 | YSVLYNSASFSTF | YSVIYNFAPFFAF | 164 | 604 |
| HLA-A*29:02 | YSVLYNSASF | YSVIYNFAPF | 1696 | 462 |
| HLA-A*03:02 | NSASFSTFK | NFAPFFAFK | 37 | 135 |
| HLA-A*30:02 | NSASFSTFKCY | NFAPFFAFKCY | 387 | 1395 |
| HLA-C*12:03 | SASFSTFKCY | FAPFFAFKCY | 498 | 139 |
| HLA-A*30:02 | VLYNSASFSTFKCY | VIYNFAPFFAFKCY | 272 | 973 |
| HLA-B*15:01 | LYNSASFSTF | IYNFAPFFAF | 468 | 1565 |
| HLA-A*68:01 | NSASFSTFK | NFAPFFAFK | 4 | 13 |
| HLA-C*03:02 | SVLYNSASF | SVIYNFAPF | 29 | 9 |
| HLA-A*11:01 | YNSASFSTFK | YNFAPFFAFK | 12 | 38 |
| HLA-A*23:01 | YSVLYNSASF | YSVIYNFAPF | 1120 | 364 |
| HLA-A*02:06 | SASFSTFKCYGV | FAPFFAFKCYGV | 1416 | 481 |
| HLA-C*03:02 | YSVLYNSASFSTF | YSVIYNFAPFFAF | 113 | 331 |
| HLA-A*29:02 | LYNSASFSTFKCY | IYNFAPFFAFKCY | 297 | 105 |
| HLA-A*68:01 | VLYNSASFSTFK | VIYNFAPFFAFK | 257 | 91 |
| HLA-A*03:02 | VLYNSASFSTFK | VIYNFAPFFAFK | 13 | 36 |
| HLA-C*14:02 | SVLYNSASF | SVIYNFAPF | 152 | 56 |
| HLA-B*15:03 | VLYNSASF | VIYNFAPF | 34 | 92 |
| HLA-B*58:01 | YSVLYNSASFSTF | YSVIYNFAPFFAF | 429 | 1153 |
| HLA-C*12:02 | SASFSTFKCY | FAPFFAFKCY | 710 | 275 |
| HLA-C*03:03 | SVLYNSASF | SVIYNFAPF | 114 | 45 |
| HLA-C*03:04 | SVLYNSASF | SVIYNFAPF | 114 | 45 |
| HLA-A*02:11 | VLYNSASFS | VIYNFAPFF | 258 | 645 |
| HLA-A*68:01 | NSASFSTFKC | NFAPFFAFKC | 281 | 702 |
| HLA-A*23:01 | DYSVLYNSASF | DYSVIYNFAPF | 1039 | 424 |
| HLA-A*33:03 | LYNSASFSTFK | IYNFAPFFAFK | 1158 | 476 |
| HLA-A*03:01 | YNSASFSTFK | YNFAPFFAFK | 80 | 194 |
| HLA-B*35:01 | YSVLYNSASF | YSVIYNFAPF | 328 | 136 |
| HLA-A*30:02 | SVLYNSASF | SVIYNFAPF | 1057 | 444 |
| HLA-A*23:01 | SVLYNSASFSTF | SVIYNFAPFFAF | 656 | 277 |
| HLA-B*15:01 | VLYNSASF | VIYNFAPF | 224 | 518 |
| HLA-B*15:01 | SASFSTFKCY | FAPFFAFKCY | 256 | 561 |
| HLA-C*03:02 | SASFSTFKCY | FAPFFAFKCY | 212 | 97 |
| HLA-C*14:02 | LYNSASFSTF | IYNFAPFFAF | 20 | 43 |
| HLA-A*68:01 | YSVLYNSASFSTFK | YSVIYNFAPFFAFK | 144 | 67 |
| HLA-A*68:01 | SVLYNSASFSTFK | SVIYNFAPFFAFK | 41 | 20 |
| HLA-A*03:01 | SVLYNSASFSTFK | SVIYNFAPFFAFK | 47 | 96 |
375-376 ST->FA
Reference VYAWNRKRISNCVADYSVLYNSASFSTFKCYGVSPTKLNDLCFTNVYADSFV
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated VYAWNRKRISNCVADYSVIYNFAPFFAFKCYGVSPTKLNDLCFTNVYADSFV
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-A*32:01 | VLYNSASFS | VIYNFAPFF | 22186 | 48 |
| HLA-A*23:01 | LYNSASFS | IYNFAPFF | 22508 | 99 |
| HLA-A*24:02 | LYNSASFS | IYNFAPFF | 25164 | 136 |
| HLA-A*23:01 | VLYNSASFS | VIYNFAPFF | 32767 | 178 |
| HLA-A*29:02 | VLYNSASFS | VIYNFAPFF | 10439 | 76 |
| HLA-C*03:02 | VLYNSASFS | VIYNFAPFF | 13004 | 100 |
| HLA-B*15:03 | VLYNSASFS | VIYNFAPFF | 3280 | 27 |
| HLA-A*26:01 | SVLYNSASFS | SVIYNFAPFF | 21993 | 202 |
| HLA-C*14:02 | LYNSASFS | IYNFAPFF | 5693 | 80 |
| HLA-A*32:01 | SVLYNSASFS | SVIYNFAPFF | 14340 | 227 |
| HLA-A*30:02 | ASFSTFKCY | APFFAFKCY | 43 | 2647 |
| HLA-A*29:02 | SVLYNSASFS | SVIYNFAPFF | 16185 | 274 |
| HLA-B*15:01 | ASFSTFKCY | APFFAFKCY | 107 | 6085 |
| HLA-B*15:01 | SVLYNSASFS | SVIYNFAPFF | 2838 | 50 |
| HLA-A*02:06 | ASFSTFKCYGV | APFFAFKCYGV | 231 | 11537 |
| HLA-A*23:01 | SVLYNSASFS | SVIYNFAPFF | 24524 | 498 |
| HLA-C*15:02 | FSTFKCYGV | FFAFKCYGV | 431 | 20837 |
| HLA-C*03:02 | SVLYNSASFS | SVIYNFAPFF | 8888 | 192 |
| HLA-C*16:01 | ASFSTFKCY | APFFAFKCY | 68 | 3117 |
| HLA-A*68:02 | SVLYNSASFST | SVIYNFAPFFA | 14147 | 319 |
| HLA-C*12:03 | ASFSTFKCY | APFFAFKCY | 115 | 5066 |
| HLA-C*16:01 | VLYNSASFS | VIYNFAPFF | 18639 | 432 |
| HLA-B*15:03 | SVLYNSASFS | SVIYNFAPFF | 2850 | 68 |
| HLA-A*23:01 | DYSVLYNSASFS | DYSVIYNFAPFF | 14260 | 341 |
| HLA-C*03:02 | ASFSTFKCY | APFFAFKCY | 64 | 2656 |
| HLA-A*11:01 | STFKCYGVSPTK | FAFKCYGVSPTK | 25 | 1030 |
| HLA-A*01:01 | NSASFSTFKCY | NFAPFFAFKCY | 328 | 13051 |
| HLA-A*68:02 | VLYNSASFST | VIYNFAPFFA | 11417 | 297 |
| HLA-C*12:03 | VLYNSASFS | VIYNFAPFF | 12383 | 333 |
| HLA-C*16:01 | NSASFSTF | NFAPFFAF | 243 | 8562 |
| HLA-A*11:01 | ASFSTFKCY | APFFAFKCY | 379 | 12606 |
| HLA-C*14:02 | VLYNSASFS | VIYNFAPFF | 11085 | 360 |
| HLA-A*23:01 | NSASFSTF | NFAPFFAF | 12240 | 424 |
| HLA-A*29:02 | YNSASFSTF | YNFAPFFAF | 1671 | 60 |
| HLA-C*16:01 | FSTFKCYGV | FFAFKCYGV | 302 | 8277 |
| HLA-B*15:01 | VLYNSASFS | VIYNFAPFF | 8189 | 317 |
| HLA-B*35:01 | ASFSTFKCY | APFFAFKCY | 2345 | 92 |
| HLA-B*58:01 | YSVLYNSASFS | YSVIYNFAPFF | 6920 | 277 |
| HLA-C*12:02 | ASFSTFKCY | APFFAFKCY | 338 | 8222 |
| HLA-A*03:01 | STFKCYGVSPTK | FAFKCYGVSPTK | 83 | 1880 |
| HLA-A*68:02 | NSASFSTFKCYGV | NFAPFFAFKCYGV | 48 | 1043 |
| HLA-A*02:06 | SVLYNSASFST | SVIYNFAPFFA | 4097 | 193 |
| HLA-C*14:02 | NSASFSTF | NFAPFFAF | 2544 | 120 |
| HLA-B*15:03 | ASFSTFKCY | APFFAFKCY | 25 | 529 |
| HLA-C*03:02 | STFKCYGVS | FAFKCYGVS | 8370 | 424 |
| HLA-B*15:01 | VLYNSASFSTF | VIYNFAPFFAF | 50 | 951 |
| HLA-A*03:02 | STFKCYGVSPTK | FAFKCYGVSPTK | 142 | 2541 |
| HLA-A*30:02 | SASFSTFKCY | FAPFFAFKCY | 71 | 1241 |
| HLA-C*14:02 | SFSTFKCY | PFFAFKCY | 386 | 6449 |
| HLA-B*15:03 | YSVLYNSASFS | YSVIYNFAPFF | 3170 | 196 |
| HLA-A*11:01 | LYNSASFSTFK | IYNFAPFFAFK | 43 | 681 |
| HLA-A*30:01 | STFKCYGVS | FAFKCYGVS | 438 | 6895 |
| HLA-A*68:02 | FSTFKCYGV | FFAFKCYGV | 17 | 261 |
| HLA-A*32:01 | YNSASFSTF | YNFAPFFAF | 4333 | 294 |
| HLA-A*02:06 | FSTFKCYGV | FFAFKCYGV | 61 | 882 |
| HLA-A*33:03 | YNSASFSTFK | YNFAPFFAFK | 823 | 57 |
| HLA-A*11:01 | FSTFKCYGVSPTK | FFAFKCYGVSPTK | 426 | 5482 |
| HLA-B*18:01 | YNSASFSTF | YNFAPFFAF | 3495 | 284 |
| HLA-A*02:06 | VLYNSASFST | VIYNFAPFFA | 710 | 58 |
| HLA-A*33:03 | NSASFSTFK | NFAPFFAFK | 181 | 16 |
| HLA-A*30:01 | LYNSASFST | IYNFAPFFA | 3467 | 316 |
| HLA-C*14:02 | FSTFKCYGV | FFAFKCYGV | 4043 | 371 |
| HLA-A*11:01 | NSASFSTFK | NFAPFFAFK | 9 | 94 |
| HLA-A*29:02 | NSASFSTFKCY | NFAPFFAFKCY | 662 | 64 |
| HLA-A*29:02 | YNSASFSTFKCY | YNFAPFFAFKCY | 842 | 86 |
| HLA-C*03:02 | YSVLYNSASFS | YSVIYNFAPFF | 2405 | 260 |
| HLA-A*29:02 | ASFSTFKCY | APFFAFKCY | 201 | 1830 |
| HLA-A*02:11 | SVLYNSASFST | SVIYNFAPFFA | 3830 | 425 |
| HLA-A*33:03 | SVLYNSASFSTFK | SVIYNFAPFFAFK | 2778 | 320 |
| HLA-A*31:01 | YNSASFSTFK | YNFAPFFAFK | 607 | 73 |
| HLA-B*35:01 | SASFSTFKCY | FAPFFAFKCY | 490 | 65 |
| HLA-A*30:01 | STFKCYGVSPTK | FAFKCYGVSPTK | 194 | 1428 |
| HLA-A*30:02 | VLYNSASFS | VIYNFAPFF | 2919 | 448 |
| HLA-A*31:01 | NSASFSTFK | NFAPFFAFK | 293 | 47 |
| HLA-B*15:03 | VLYNSASFSTF | VIYNFAPFFAF | 38 | 235 |
| HLA-A*29:02 | LYNSASFSTF | IYNFAPFFAF | 387 | 63 |
| HLA-A*31:01 | SVLYNSASFSTFK | SVIYNFAPFFAFK | 857 | 150 |
| HLA-A*02:01 | VLYNSASFST | VIYNFAPFFA | 243 | 43 |
| HLA-A*23:01 | YNSASFSTF | YNFAPFFAF | 352 | 63 |
| HLA-A*03:01 | VLYNSASFSTFKC | VIYNFAPFFAFKC | 312 | 1619 |
| HLA-A*03:01 | LYNSASFSTFK | IYNFAPFFAFK | 295 | 1419 |
| HLA-A*03:01 | NSASFSTFK | NFAPFFAFK | 113 | 494 |
| HLA-B*35:01 | YNSASFSTF | YNFAPFFAF | 760 | 180 |
| HLA-A*02:02 | VLYNSASFST | VIYNFAPFFA | 202 | 49 |
| HLA-A*31:01 | VLYNSASFSTFK | VIYNFAPFFAFK | 565 | 139 |
| HLA-A*01:01 | SASFSTFKCY | FAPFFAFKCY | 461 | 1849 |
| HLA-A*11:01 | SASFSTFK | FAPFFAFK | 452 | 1795 |
| HLA-A*03:01 | VLYNSASFSTFK | VIYNFAPFFAFK | 7 | 27 |
| HLA-A*02:11 | VLYNSASFST | VIYNFAPFFA | 77 | 20 |
| HLA-A*68:01 | SASFSTFK | FAPFFAFK | 1114 | 290 |
| HLA-A*03:02 | LYNSASFSTFK | IYNFAPFFAFK | 202 | 760 |
| HLA-A*31:01 | LYNSASFSTFK | IYNFAPFFAFK | 299 | 81 |
| HLA-B*15:01 | YSVLYNSASFSTF | YSVIYNFAPFFAF | 164 | 604 |
| HLA-A*03:02 | NSASFSTFK | NFAPFFAFK | 37 | 135 |
| HLA-A*30:02 | NSASFSTFKCY | NFAPFFAFKCY | 387 | 1395 |
| HLA-C*12:03 | SASFSTFKCY | FAPFFAFKCY | 498 | 139 |
| HLA-A*30:02 | VLYNSASFSTFKCY | VIYNFAPFFAFKCY | 272 | 973 |
| HLA-B*15:01 | LYNSASFSTF | IYNFAPFFAF | 468 | 1565 |
| HLA-A*68:01 | NSASFSTFK | NFAPFFAFK | 4 | 13 |
| HLA-A*11:01 | YNSASFSTFK | YNFAPFFAFK | 12 | 38 |
| HLA-A*02:06 | SASFSTFKCYGV | FAPFFAFKCYGV | 1416 | 481 |
| HLA-C*03:02 | YSVLYNSASFSTF | YSVIYNFAPFFAF | 113 | 331 |
| HLA-A*68:01 | FSTFKCYGVSPTK | FFAFKCYGVSPTK | 288 | 832 |
| HLA-A*29:02 | LYNSASFSTFKCY | IYNFAPFFAFKCY | 297 | 105 |
| HLA-A*68:01 | VLYNSASFSTFK | VIYNFAPFFAFK | 257 | 91 |
| HLA-A*03:02 | VLYNSASFSTFK | VIYNFAPFFAFK | 13 | 36 |
| HLA-B*58:01 | YSVLYNSASFSTF | YSVIYNFAPFFAF | 429 | 1153 |
| HLA-C*12:02 | SASFSTFKCY | FAPFFAFKCY | 710 | 275 |
| HLA-A*02:11 | VLYNSASFS | VIYNFAPFF | 258 | 645 |
| HLA-A*68:01 | NSASFSTFKC | NFAPFFAFKC | 281 | 702 |
| HLA-A*33:03 | LYNSASFSTFK | IYNFAPFFAFK | 1158 | 476 |
| HLA-A*03:01 | YNSASFSTFK | YNFAPFFAFK | 80 | 194 |
| HLA-A*23:01 | SVLYNSASFSTF | SVIYNFAPFFAF | 656 | 277 |
| HLA-B*15:01 | SASFSTFKCY | FAPFFAFKCY | 256 | 561 |
| HLA-C*03:02 | SASFSTFKCY | FAPFFAFKCY | 212 | 97 |
| HLA-C*14:02 | LYNSASFSTF | IYNFAPFFAF | 20 | 43 |
| HLA-A*68:01 | YSVLYNSASFSTFK | YSVIYNFAPFFAFK | 144 | 67 |
| HLA-A*30:01 | TFKCYGVSPTK | AFKCYGVSPTK | 319 | 155 |
| HLA-A*68:01 | SVLYNSASFSTFK | SVIYNFAPFFAFK | 41 | 20 |
| HLA-A*03:01 | SVLYNSASFSTFK | SVIYNFAPFFAFK | 47 | 96 |
| HLA-A*02:01 | FSTFKCYGV | FFAFKCYGV | 457 | 926 |
D405N
Reference YGVSPTKLNDLCFTNVYADSFVIRGDEVRQIAPGQTGKIADYNYKLPDDFT
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated YGVSPTKLNDLCFTNVYADSFVIRGNEVSQIAPGQTGNIADYNYKLPDDFT
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-A*68:01 | FVIRGDEVR | FVIRGNEVS | 23 | 15265 |
| HLA-A*68:01 | SFVIRGDEVR | SFVIRGNEVS | 186 | 29639 |
| HLA-A*68:01 | DSFVIRGDEVR | DSFVIRGNEVS | 220 | 29286 |
| HLA-A*33:03 | FVIRGDEVR | FVIRGNEVS | 320 | 29448 |
| HLA-A*33:03 | SFVIRGDEVR | SFVIRGNEVS | 459 | 31986 |
| HLA-C*14:02 | SFVIRGDEV | SFVIRGNEV | 216 | 46 |
| HLA-C*06:02 | IRGDEVRQI | IRGNEVSQI | 147 | 642 |
R408S
Reference SPTKLNDLCFTNVYADSFVIRGDEVRQIAPGQTGKIADYNYKLPDDFTGCV
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated SPTKLNDLCFTNVYADSFVIRGNEVSQIAPGQTGNIADYNYKLPDDFTGCV
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-A*68:01 | FVIRGDEVR | FVIRGNEVS | 23 | 15265 |
| HLA-A*03:01 | RQIAPGQTGK | SQIAPGQTGN | 49 | 32429 |
| HLA-A*11:01 | RQIAPGQTGK | SQIAPGQTGN | 103 | 32387 |
| HLA-A*03:02 | RQIAPGQTGK | SQIAPGQTGN | 123 | 32767 |
| HLA-A*68:01 | EVRQIAPGQTGK | EVSQIAPGQTGN | 144 | 32361 |
| HLA-A*68:01 | SFVIRGDEVR | SFVIRGNEVS | 186 | 29639 |
| HLA-A*68:01 | DSFVIRGDEVR | DSFVIRGNEVS | 220 | 29286 |
| HLA-A*33:03 | FVIRGDEVR | FVIRGNEVS | 320 | 29448 |
| HLA-A*33:03 | SFVIRGDEVR | SFVIRGNEVS | 459 | 31986 |
| HLA-A*30:01 | RQIAPGQTGK | SQIAPGQTGN | 275 | 18394 |
| HLA-C*06:02 | IRGDEVRQI | IRGNEVSQI | 147 | 642 |
| HLA-B*15:03 | RQIAPGQT | SQIAPGQT | 478 | 1764 |
| HLA-B*15:03 | RQIAPGQTGKIADY | SQIAPGQTGNIADY | 17 | 55 |
| HLA-A*30:02 | RQIAPGQTGKIADY | SQIAPGQTGNIADY | 324 | 951 |
| HLA-B*15:03 | RQIAPGQTGKI | SQIAPGQTGNI | 236 | 691 |
| HLA-B*15:03 | RQIAPGQTG | SQIAPGQTG | 39 | 107 |
K417N
Reference FTNVYADSFVIRGDEVRQIAPGQTGKIADYNYKLPDDFTGCVIAWNSNNLD
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated FTNVYADSFVIRGNEVSQIAPGQTGNIADYNYKLPDDFTGCVIAWNSNKLD
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-A*03:01 | RQIAPGQTGK | SQIAPGQTGN | 49 | 32429 |
| HLA-A*11:01 | RQIAPGQTGK | SQIAPGQTGN | 103 | 32387 |
| HLA-A*03:02 | RQIAPGQTGK | SQIAPGQTGN | 123 | 32767 |
| HLA-A*68:01 | EVRQIAPGQTGK | EVSQIAPGQTGN | 144 | 32361 |
| HLA-A*68:01 | QIAPGQTGK | QIAPGQTGN | 136 | 27997 |
| HLA-A*11:01 | QIAPGQTGK | QIAPGQTGN | 165 | 32767 |
| HLA-A*03:02 | QIAPGQTGK | QIAPGQTGN | 278 | 32767 |
| HLA-A*03:01 | QIAPGQTGK | QIAPGQTGN | 296 | 32767 |
| HLA-A*32:01 | KIADYNYKL | NIADYNYKL | 58 | 4480 |
| HLA-A*30:01 | RQIAPGQTGK | SQIAPGQTGN | 275 | 18394 |
| HLA-A*68:02 | KIADYNYKL | NIADYNYKL | 1274 | 39 |
| HLA-A*03:02 | KIADYNYK | NIADYNYK | 476 | 6850 |
| HLA-A*02:06 | KIADYNYKL | NIADYNYKL | 20 | 101 |
| HLA-A*02:01 | KIADYNYKL | NIADYNYKL | 23 | 110 |
| HLA-C*17:01 | KIADYNYKL | NIADYNYKL | 155 | 669 |
| HLA-A*30:01 | TGKIADYNYK | TGNIADYNYK | 292 | 1164 |
| HLA-A*02:11 | KIADYNYKL | NIADYNYKL | 5 | 19 |
| HLA-B*15:03 | RQIAPGQTGKIADY | SQIAPGQTGNIADY | 17 | 55 |
| HLA-A*30:02 | RQIAPGQTGKIADY | SQIAPGQTGNIADY | 324 | 951 |
| HLA-B*15:03 | RQIAPGQTGKI | SQIAPGQTGNI | 236 | 691 |
| HLA-A*68:01 | TGKIADYNYK | TGNIADYNYK | 957 | 343 |
| HLA-A*01:01 | QTGKIADYNY | QTGNIADYNY | 580 | 270 |
N440K
Reference TGKIADYNYKLPDDFTGCVIAWNSNNLDSKVGGNYNYLYRLFRKSNLKPFE
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated TGNIADYNYKLPDDFTGCVIAWNSNKLDSKPSGNYNYLYRLFRKSKLKPFE
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-A*11:01 | CVIAWNSNN | CVIAWNSNK | 24486 | 26 |
| HLA-A*68:01 | CVIAWNSNN | CVIAWNSNK | 11802 | 16 |
| HLA-A*68:01 | FTGCVIAWNSNN | FTGCVIAWNSNK | 25655 | 59 |
| HLA-A*03:01 | CVIAWNSNN | CVIAWNSNK | 24643 | 61 |
| HLA-A*03:02 | CVIAWNSNN | CVIAWNSNK | 22621 | 61 |
| HLA-A*11:01 | GCVIAWNSNN | GCVIAWNSNK | 32767 | 94 |
| HLA-A*03:01 | GCVIAWNSNN | GCVIAWNSNK | 32767 | 128 |
| HLA-A*11:01 | TGCVIAWNSNN | TGCVIAWNSNK | 32767 | 153 |
| HLA-A*68:01 | TGCVIAWNSNN | TGCVIAWNSNK | 32767 | 180 |
| HLA-A*68:01 | DFTGCVIAWNSNN | DFTGCVIAWNSNK | 32767 | 255 |
| HLA-A*03:02 | GCVIAWNSNN | GCVIAWNSNK | 32767 | 277 |
| HLA-A*11:01 | FTGCVIAWNSNN | FTGCVIAWNSNK | 32767 | 408 |
| HLA-A*31:01 | CVIAWNSNN | CVIAWNSNK | 18631 | 279 |
| HLA-A*33:03 | CVIAWNSNN | CVIAWNSNK | 20952 | 493 |
| HLA-A*30:01 | CVIAWNSNN | CVIAWNSNK | 11396 | 467 |
| HLA-A*30:02 | NLDSKVGGNYNYLY | KLDSKPSGNYNYLY | 3335 | 366 |
| HLA-A*02:02 | NLDSKVGGNYNYL | KLDSKPSGNYNYL | 1730 | 219 |
| HLA-A*02:11 | NLDSKVGGNYNYL | KLDSKPSGNYNYL | 671 | 104 |
| HLA-A*30:02 | NLDSKVGGNY | KLDSKPSGNY | 422 | 69 |
| HLA-B*15:01 | NLDSKVGGNY | KLDSKPSGNY | 1404 | 287 |
| HLA-A*01:01 | NLDSKVGGNY | KLDSKPSGNY | 371 | 887 |
| HLA-A*01:01 | NLDSKVGGNYNYLY | KLDSKPSGNYNYLY | 193 | 427 |
| HLA-A*02:11 | VIAWNSNNL | VIAWNSNKL | 269 | 586 |
| HLA-A*68:02 | FTGCVIAWNSNNL | FTGCVIAWNSNKL | 461 | 960 |
445-446 VG->PS
Reference DYNYKLPDDFTGCVIAWNSNNLDSKVGGNYNYLYRLFRKSNLKPFERDISTE
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated DYNYKLPDDFTGCVIAWNSNKLDSKPSGNYNYLYRLFRKSKLKPFERDISTE
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-B*07:02 | KVGGNYNYL | KPSGNYNYL | 12674 | 46 |
| HLA-B*42:01 | KVGGNYNYL | KPSGNYNYL | 7890 | 37 |
| HLA-A*02:11 | KVGGNYNYL | KPSGNYNYL | 111 | 9769 |
| HLA-B*42:01 | SKVGGNYNYL | SKPSGNYNYL | 18106 | 267 |
| HLA-A*02:11 | KVGGNYNYLYRL | KPSGNYNYLYRL | 274 | 16428 |
| HLA-B*07:02 | SKVGGNYNYL | SKPSGNYNYL | 23912 | 402 |
| HLA-A*02:02 | KVGGNYNYL | KPSGNYNYL | 279 | 10673 |
| HLA-A*02:06 | KVGGNYNYL | KPSGNYNYL | 452 | 12158 |
| HLA-A*30:02 | KVGGNYNY | KPSGNYNY | 420 | 6251 |
| HLA-A*01:01 | VGGNYNYLY | PSGNYNYLY | 2993 | 213 |
| HLA-B*15:03 | SKVGGNYNY | SKPSGNYNY | 25 | 286 |
| HLA-A*31:01 | KVGGNYNYLYRLFR | KPSGNYNYLYRLFR | 36 | 337 |
| HLA-A*30:02 | NLDSKVGGNYNYLY | KLDSKPSGNYNYLY | 3335 | 366 |
| HLA-A*02:02 | NLDSKVGGNYNYL | KLDSKPSGNYNYL | 1730 | 219 |
| HLA-A*31:01 | KVGGNYNYLYR | KPSGNYNYLYR | 66 | 491 |
| HLA-A*30:02 | VGGNYNYLY | PSGNYNYLY | 45 | 291 |
| HLA-A*02:11 | NLDSKVGGNYNYL | KLDSKPSGNYNYL | 671 | 104 |
| HLA-B*58:01 | KVGGNYNYLY | KPSGNYNYLY | 1217 | 190 |
| HLA-A*30:02 | NLDSKVGGNY | KLDSKPSGNY | 422 | 69 |
| HLA-B*15:03 | SKVGGNYNYLY | SKPSGNYNYLY | 313 | 1847 |
| HLA-A*24:02 | GGNYNYLYRLF | SGNYNYLYRLF | 1816 | 313 |
| HLA-A*23:01 | VGGNYNYLYRLF | PSGNYNYLYRLF | 222 | 1273 |
| HLA-A*68:01 | GGNYNYLYR | SGNYNYLYR | 1245 | 229 |
| HLA-A*23:01 | GGNYNYLYRLF | SGNYNYLYRLF | 988 | 186 |
| HLA-A*33:03 | GGNYNYLYR | SGNYNYLYR | 1064 | 201 |
| HLA-B*15:01 | NLDSKVGGNY | KLDSKPSGNY | 1404 | 287 |
| HLA-A*29:02 | VGGNYNYLY | PSGNYNYLY | 94 | 438 |
| HLA-A*24:02 | VGGNYNYLYRLF | PSGNYNYLYRLF | 391 | 1786 |
| HLA-A*31:01 | VGGNYNYLYR | PSGNYNYLYR | 138 | 585 |
| HLA-A*29:02 | KVGGNYNYLY | KPSGNYNYLY | 100 | 414 |
| HLA-A*31:01 | VGGNYNYLYRLFR | PSGNYNYLYRLFR | 253 | 739 |
| HLA-A*31:01 | GGNYNYLYRLFR | SGNYNYLYRLFR | 153 | 56 |
| HLA-A*33:03 | GGNYNYLYRLFR | SGNYNYLYRLFR | 636 | 251 |
| HLA-A*01:01 | NLDSKVGGNY | KLDSKPSGNY | 371 | 887 |
| HLA-A*33:03 | DSKVGGNYNYLYR | DSKPSGNYNYLYR | 528 | 224 |
| HLA-A*11:01 | GGNYNYLYR | SGNYNYLYR | 876 | 391 |
| HLA-A*01:01 | NLDSKVGGNYNYLY | KLDSKPSGNYNYLY | 193 | 427 |
N460K
Reference AWNSNNLDSKVGGNYNYLYRLFRKSNLKPFERDISTEIYQAGSTPCNGVEG
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated AWNSNKLDSKPSGNYNYLYRLFRKSKLKPFERDISTEIYQAGNRPCNGVAG
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-A*03:01 | YLYRLFRKSN | YLYRLFRKSK | 23374 | 37 |
| HLA-A*03:02 | YLYRLFRKSN | YLYRLFRKSK | 28215 | 198 |
| HLA-A*31:01 | LYRLFRKSN | LYRLFRKSK | 21575 | 190 |
| HLA-A*30:01 | LYRLFRKSN | LYRLFRKSK | 1444 | 30 |
| HLA-A*31:01 | YLYRLFRKSN | YLYRLFRKSK | 21799 | 460 |
| HLA-A*30:01 | YLYRLFRKSN | YLYRLFRKSK | 5606 | 156 |
| HLA-A*11:01 | KSNLKPFER | KSKLKPFER | 401 | 2898 |
| HLA-B*08:01 | FRKSNLKPF | FRKSKLKPF | 777 | 126 |
| HLA-A*68:01 | YRLFRKSNLK | YRLFRKSKLK | 353 | 1913 |
| HLA-A*03:02 | RLFRKSNLKP | RLFRKSKLKP | 220 | 1183 |
| HLA-B*15:03 | FRKSNLKPF | FRKSKLKPF | 72 | 372 |
| HLA-A*03:02 | LYRLFRKSNLK | LYRLFRKSKLK | 242 | 1236 |
| HLA-A*11:01 | YRLFRKSNLK | YRLFRKSKLK | 170 | 837 |
| HLA-A*30:01 | KSNLKPFER | KSKLKPFER | 964 | 213 |
| HLA-A*03:02 | YRLFRKSNLK | YRLFRKSKLK | 44 | 186 |
| HLA-B*15:03 | RLFRKSNLKPF | RLFRKSKLKPF | 56 | 224 |
| HLA-A*11:01 | RLFRKSNLK | RLFRKSKLK | 24 | 95 |
| HLA-B*15:01 | RLFRKSNLKPF | RLFRKSKLKPF | 113 | 395 |
| HLA-B*15:03 | LFRKSNLKPF | LFRKSKLKPF | 393 | 1267 |
| HLA-A*03:02 | YLYRLFRKSNLK | YLYRLFRKSKLK | 134 | 416 |
| HLA-B*15:03 | YRLFRKSNLKPF | YRLFRKSKLKPF | 454 | 1357 |
| HLA-A*32:01 | RLFRKSNLKPF | RLFRKSKLKPF | 121 | 361 |
| HLA-A*03:01 | RLFRKSNLKPF | RLFRKSKLKPF | 439 | 1261 |
| HLA-B*15:03 | RKSNLKPF | RKSKLKPF | 191 | 546 |
| HLA-C*14:02 | LFRKSNLKPF | LFRKSKLKPF | 252 | 673 |
| HLA-A*03:01 | RLFRKSNLKP | RLFRKSKLKP | 32 | 82 |
| HLA-C*14:02 | FRKSNLKPF | FRKSKLKPF | 232 | 584 |
| HLA-C*07:02 | FRKSNLKPF | FRKSKLKPF | 135 | 324 |
| HLA-A*03:02 | RLFRKSNLK | RLFRKSKLK | 8 | 19 |
| HLA-A*31:01 | RLFRKSNLK | RLFRKSKLK | 25 | 59 |
| HLA-A*31:01 | LYRLFRKSNLK | LYRLFRKSKLK | 356 | 840 |
| HLA-A*03:01 | LYRLFRKSNLK | LYRLFRKSKLK | 65 | 146 |
| HLA-A*74:01 | RLFRKSNLK | RLFRKSKLK | 77 | 166 |
477-478 ST->NR
Reference LYRLFRKSNLKPFERDISTEIYQAGSTPCNGVEGFNCYFPLQSYGFQPTNGV
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated LYRLFRKSKLKPFERDISTEIYQAGNRPCNGVAGPNCYSPLQSYGFRPTYGV
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-A*68:01 | STEIYQAGST | STEIYQAGNR | 22142 | 35 |
| HLA-B*15:03 | YQAGSTPCNGVEGF | YQAGNRPCNGVAGP | 217 | 25822 |
| HLA-B*35:01 | TPCNGVEGF | RPCNGVAGP | 319 | 26613 |
| HLA-A*33:03 | STEIYQAGST | STEIYQAGNR | 32767 | 483 |
| HLA-B*15:01 | YQAGSTPCNGVEGF | YQAGNRPCNGVAGP | 408 | 26793 |
| HLA-B*50:01 | TEIYQAGSTP | TEIYQAGNRP | 464 | 1936 |
| HLA-A*02:11 | YQAGSTPCNGV | YQAGNRPCNGV | 173 | 656 |
| HLA-A*02:06 | YQAGSTPCNGV | YQAGNRPCNGV | 39 | 144 |
| HLA-A*02:01 | YQAGSTPCNGV | YQAGNRPCNGV | 420 | 1481 |
| HLA-A*02:02 | YQAGSTPCNGV | YQAGNRPCNGV | 107 | 230 |
E484A
Reference SNLKPFERDISTEIYQAGSTPCNGVEGFNCYFPLQSYGFQPTNGVGYQPYR
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated SKLKPFERDISTEIYQAGNRPCNGVAGPNCYSPLQSYGFRPTYGVGHQPYR
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-B*40:02 | VEGFNCYFPL | VAGPNCYSPL | 31 | 28153 |
| HLA-B*40:01 | VEGFNCYFPL | VAGPNCYSPL | 76 | 29734 |
| HLA-C*01:02 | EGFNCYFPL | AGPNCYSPL | 14564 | 97 |
| HLA-B*15:03 | YQAGSTPCNGVEGF | YQAGNRPCNGVAGP | 217 | 25822 |
| HLA-C*01:02 | VEGFNCYFPL | VAGPNCYSPL | 20165 | 183 |
| HLA-A*68:02 | EGFNCYFPL | AGPNCYSPL | 117 | 11181 |
| HLA-B*35:01 | TPCNGVEGF | RPCNGVAGP | 319 | 26613 |
| HLA-B*15:01 | YQAGSTPCNGVEGF | YQAGNRPCNGVAGP | 408 | 26793 |
| HLA-C*03:02 | VEGFNCYFPLQSY | VAGPNCYSPLQSY | 14989 | 314 |
| HLA-A*02:11 | GVEGFNCYFPL | GVAGPNCYSPL | 464 | 2693 |
| HLA-B*35:01 | NGVEGFNCY | NGVAGPNCY | 346 | 1951 |
| HLA-A*02:06 | GVEGFNCYFPL | GVAGPNCYSPL | 400 | 2162 |
F486P
Reference LKPFERDISTEIYQAGSTPCNGVEGFNCYFPLQSYGFQPTNGVGYQPYRVV
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated LKPFERDISTEIYQAGNRPCNGVAGPNCYSPLQSYGFRPTYGVGHQPYRVV
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-B*40:02 | VEGFNCYFPL | VAGPNCYSPL | 31 | 28153 |
| HLA-B*40:01 | VEGFNCYFPL | VAGPNCYSPL | 76 | 29734 |
| HLA-B*07:02 | GFNCYFPL | GPNCYSPL | 27420 | 143 |
| HLA-C*01:02 | EGFNCYFPL | AGPNCYSPL | 14564 | 97 |
| HLA-B*15:03 | YQAGSTPCNGVEGF | YQAGNRPCNGVAGP | 217 | 25822 |
| HLA-C*01:02 | VEGFNCYFPL | VAGPNCYSPL | 20165 | 183 |
| HLA-B*42:01 | GFNCYFPL | GPNCYSPL | 15642 | 154 |
| HLA-A*68:02 | EGFNCYFPL | AGPNCYSPL | 117 | 11181 |
| HLA-B*35:01 | TPCNGVEGF | RPCNGVAGP | 319 | 26613 |
| HLA-B*15:01 | YQAGSTPCNGVEGF | YQAGNRPCNGVAGP | 408 | 26793 |
| HLA-A*29:02 | FNCYFPLQSY | PNCYSPLQSY | 73 | 4504 |
| HLA-B*15:03 | FNCYFPLQSY | PNCYSPLQSY | 101 | 6104 |
| HLA-C*03:02 | FNCYFPLQSY | PNCYSPLQSY | 225 | 12092 |
| HLA-C*03:02 | VEGFNCYFPLQSY | VAGPNCYSPLQSY | 14989 | 314 |
| HLA-A*29:02 | GFNCYFPLQSY | GPNCYSPLQSY | 191 | 7026 |
| HLA-A*02:11 | GVEGFNCYFPL | GVAGPNCYSPL | 464 | 2693 |
| HLA-B*35:01 | NGVEGFNCY | NGVAGPNCY | 346 | 1951 |
| HLA-A*02:06 | GVEGFNCYFPL | GVAGPNCYSPL | 400 | 2162 |
| HLA-A*30:02 | FNCYFPLQSY | PNCYSPLQSY | 425 | 2051 |
F490S
Reference ERDISTEIYQAGSTPCNGVEGFNCYFPLQSYGFQPTNGVGYQPYRVVVLSF
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated ERDISTEIYQAGNRPCNGVAGPNCYSPLQSYGFRPTYGVGHQPYRVVVLSF
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-B*40:02 | VEGFNCYFPL | VAGPNCYSPL | 31 | 28153 |
| HLA-B*40:01 | VEGFNCYFPL | VAGPNCYSPL | 76 | 29734 |
| HLA-A*68:01 | YFPLQSYGFQ | YSPLQSYGFR | 14480 | 50 |
| HLA-B*07:02 | GFNCYFPL | GPNCYSPL | 27420 | 143 |
| HLA-C*03:02 | YFPLQSYGFQPTN | YSPLQSYGFRPTY | 27639 | 179 |
| HLA-C*01:02 | EGFNCYFPL | AGPNCYSPL | 14564 | 97 |
| HLA-A*31:01 | YFPLQSYGFQ | YSPLQSYGFR | 16290 | 110 |
| HLA-B*15:03 | FPLQSYGFQPTN | SPLQSYGFRPTY | 20159 | 178 |
| HLA-B*15:03 | YFPLQSYGFQPTN | YSPLQSYGFRPTY | 22168 | 197 |
| HLA-A*33:03 | YFPLQSYGFQ | YSPLQSYGFR | 11148 | 101 |
| HLA-C*01:02 | VEGFNCYFPL | VAGPNCYSPL | 20165 | 183 |
| HLA-A*30:02 | FPLQSYGFQPTN | SPLQSYGFRPTY | 21475 | 197 |
| HLA-C*16:01 | YFPLQSYGFQPTN | YSPLQSYGFRPTY | 29358 | 276 |
| HLA-B*42:01 | GFNCYFPL | GPNCYSPL | 15642 | 154 |
| HLA-A*33:03 | FPLQSYGFQ | SPLQSYGFR | 23527 | 238 |
| HLA-A*68:02 | EGFNCYFPL | AGPNCYSPL | 117 | 11181 |
| HLA-A*31:01 | CYFPLQSYGFQ | CYSPLQSYGFR | 8535 | 105 |
| HLA-A*33:03 | CYFPLQSYGFQ | CYSPLQSYGFR | 11311 | 156 |
| HLA-A*29:02 | FNCYFPLQSY | PNCYSPLQSY | 73 | 4504 |
| HLA-B*15:03 | FNCYFPLQSY | PNCYSPLQSY | 101 | 6104 |
| HLA-C*03:02 | FNCYFPLQSY | PNCYSPLQSY | 225 | 12092 |
| HLA-A*33:03 | NCYFPLQSYGFQ | NCYSPLQSYGFR | 22134 | 435 |
| HLA-A*30:02 | YFPLQSYGFQPTN | YSPLQSYGFRPTY | 18743 | 373 |
| HLA-A*29:02 | YFPLQSYGFQPTN | YSPLQSYGFRPTY | 19514 | 389 |
| HLA-C*03:02 | VEGFNCYFPLQSY | VAGPNCYSPLQSY | 14989 | 314 |
| HLA-A*29:02 | GFNCYFPLQSY | GPNCYSPLQSY | 191 | 7026 |
| HLA-C*14:02 | YFPLQSYGF | YSPLQSYGF | 18 | 646 |
| HLA-B*35:01 | FPLQSYGF | SPLQSYGF | 197 | 6313 |
| HLA-B*53:01 | FPLQSYGF | SPLQSYGF | 323 | 8460 |
| HLA-A*29:02 | YFPLQSYGF | YSPLQSYGF | 213 | 4114 |
| HLA-A*23:01 | YFPLQSYGF | YSPLQSYGF | 49 | 724 |
| HLA-C*16:01 | YFPLQSYGF | YSPLQSYGF | 4518 | 311 |
| HLA-A*24:02 | YFPLQSYGF | YSPLQSYGF | 50 | 655 |
| HLA-C*03:02 | YFPLQSYGF | YSPLQSYGF | 3990 | 334 |
| HLA-C*01:02 | YFPLQSYGF | YSPLQSYGF | 2910 | 404 |
| HLA-A*02:11 | GVEGFNCYFPL | GVAGPNCYSPL | 464 | 2693 |
| HLA-A*02:06 | GVEGFNCYFPL | GVAGPNCYSPL | 400 | 2162 |
| HLA-C*07:02 | YFPLQSYGF | YSPLQSYGF | 447 | 2163 |
| HLA-A*30:02 | FNCYFPLQSY | PNCYSPLQSY | 425 | 2051 |
| HLA-A*29:02 | CYFPLQSYGF | CYSPLQSYGF | 93 | 444 |
| HLA-A*23:01 | NCYFPLQSYGF | NCYSPLQSYGF | 271 | 1221 |
| HLA-A*23:01 | CYFPLQSYGFQ | CYSPLQSYGFR | 498 | 1935 |
| HLA-A*23:01 | CYFPLQSYGF | CYSPLQSYGF | 10 | 28 |
Q498R
Reference YQAGSTPCNGVEGFNCYFPLQSYGFQPTNGVGYQPYRVVVLSFELLHAPAT
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated YQAGNRPCNGVAGPNCYSPLQSYGFRPTYGVGHQPYRVVVLSFELLHAPAT
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-C*03:02 | QSYGFQPTN | QSYGFRPTY | 12903 | 18 |
| HLA-B*15:01 | LQSYGFQPTN | LQSYGFRPTY | 6135 | 9 |
| HLA-C*16:01 | QSYGFQPTN | QSYGFRPTY | 10868 | 16 |
| HLA-A*29:02 | QSYGFQPTN | QSYGFRPTY | 26152 | 43 |
| HLA-B*15:03 | QSYGFQPTN | QSYGFRPTY | 6543 | 11 |
| HLA-B*15:03 | LQSYGFQPTN | LQSYGFRPTY | 2045 | 4 |
| HLA-C*12:03 | QSYGFQPTN | QSYGFRPTY | 12696 | 25 |
| HLA-B*15:03 | PLQSYGFQPTN | PLQSYGFRPTY | 22802 | 60 |
| HLA-C*14:02 | SYGFQPTN | SYGFRPTY | 20230 | 66 |
| HLA-A*68:01 | YFPLQSYGFQ | YSPLQSYGFR | 14480 | 50 |
| HLA-C*03:02 | LQSYGFQPTN | LQSYGFRPTY | 23421 | 125 |
| HLA-B*15:01 | FQPTNGVGY | FRPTYGVGH | 166 | 29043 |
| HLA-A*30:02 | QSYGFQPTN | QSYGFRPTY | 5297 | 32 |
| HLA-B*35:01 | QSYGFQPTN | QSYGFRPTY | 24615 | 156 |
| HLA-C*03:02 | YFPLQSYGFQPTN | YSPLQSYGFRPTY | 27639 | 179 |
| HLA-A*31:01 | YFPLQSYGFQ | YSPLQSYGFR | 16290 | 110 |
| HLA-C*16:01 | LQSYGFQPTN | LQSYGFRPTY | 27149 | 187 |
| HLA-A*30:02 | LQSYGFQPTN | LQSYGFRPTY | 7508 | 54 |
| HLA-B*15:01 | QSYGFQPTN | QSYGFRPTY | 21529 | 157 |
| HLA-C*12:02 | QSYGFQPTN | QSYGFRPTY | 21029 | 169 |
| HLA-B*15:03 | FPLQSYGFQPTN | SPLQSYGFRPTY | 20159 | 178 |
| HLA-B*15:03 | YFPLQSYGFQPTN | YSPLQSYGFRPTY | 22168 | 197 |
| HLA-A*33:03 | YFPLQSYGFQ | YSPLQSYGFR | 11148 | 101 |
| HLA-A*30:02 | FPLQSYGFQPTN | SPLQSYGFRPTY | 21475 | 197 |
| HLA-A*29:02 | LQSYGFQPTN | LQSYGFRPTY | 28458 | 265 |
| HLA-C*16:01 | YFPLQSYGFQPTN | YSPLQSYGFRPTY | 29358 | 276 |
| HLA-B*15:03 | FQPTNGVGY | FRPTYGVGH | 63 | 6390 |
| HLA-A*29:02 | GFQPTNGVGY | GFRPTYGVGH | 188 | 18753 |
| HLA-A*33:03 | FPLQSYGFQ | SPLQSYGFR | 23527 | 238 |
| HLA-C*14:02 | QSYGFQPTN | QSYGFRPTY | 25941 | 277 |
| HLA-C*12:03 | LQSYGFQPTN | LQSYGFRPTY | 26641 | 325 |
| HLA-A*31:01 | CYFPLQSYGFQ | CYSPLQSYGFR | 8535 | 105 |
| HLA-B*15:03 | LQSYGFQPTNG | LQSYGFRPTYG | 6387 | 82 |
| HLA-A*32:01 | QSYGFQPTN | QSYGFRPTY | 16619 | 226 |
| HLA-A*11:01 | QSYGFQPTN | QSYGFRPTY | 22068 | 301 |
| HLA-A*33:03 | CYFPLQSYGFQ | CYSPLQSYGFR | 11311 | 156 |
| HLA-C*02:02 | QSYGFQPTN | QSYGFRPTY | 26598 | 412 |
| HLA-C*02:10 | QSYGFQPTN | QSYGFRPTY | 26598 | 412 |
| HLA-A*33:03 | NCYFPLQSYGFQ | NCYSPLQSYGFR | 22134 | 435 |
| HLA-A*30:02 | YFPLQSYGFQPTN | YSPLQSYGFRPTY | 18743 | 373 |
| HLA-A*29:02 | YFPLQSYGFQPTN | YSPLQSYGFRPTY | 19514 | 389 |
| HLA-B*15:01 | LQSYGFQPTNGVGY | LQSYGFRPTYGVGH | 180 | 8335 |
| HLA-B*15:01 | FQPTNGVGYQPY | FRPTYGVGHQPY | 290 | 11634 |
| HLA-B*15:03 | YGFQPTNGVGY | YGFRPTYGVGH | 368 | 10641 |
| HLA-A*30:02 | GFQPTNGVGY | GFRPTYGVGH | 169 | 4730 |
| HLA-A*30:02 | QSYGFQPTNGVGY | QSYGFRPTYGVGH | 258 | 7114 |
| HLA-B*15:03 | LQSYGFQPTNGVGY | LQSYGFRPTYGVGH | 113 | 2917 |
| HLA-C*07:02 | FQPTNGVGYQPY | FRPTYGVGHQPY | 6169 | 356 |
| HLA-B*57:01 | QSYGFQPTN | QSYGFRPTY | 4070 | 293 |
| HLA-A*30:01 | QSYGFQPTN | QSYGFRPTY | 3720 | 308 |
| HLA-B*57:01 | LQSYGFQPTN | LQSYGFRPTY | 3839 | 375 |
| HLA-C*14:02 | FQPTNGVGYQPY | FRPTYGVGHQPY | 3107 | 366 |
| HLA-B*15:03 | FQPTNGVGYQPY | FRPTYGVGHQPY | 183 | 1236 |
| HLA-A*31:01 | QPTNGVGYQPYR | RPTYGVGHQPYR | 3100 | 478 |
| HLA-B*58:01 | QSYGFQPTN | QSYGFRPTY | 1532 | 249 |
| HLA-A*23:01 | SYGFQPTNGV | SYGFRPTYGV | 2697 | 483 |
| HLA-A*02:11 | YGFQPTNGV | YGFRPTYGV | 307 | 59 |
| HLA-B*15:03 | LQSYGFQPTNGV | LQSYGFRPTYGV | 1971 | 442 |
| HLA-A*23:01 | CYFPLQSYGFQ | CYSPLQSYGFR | 498 | 1935 |
| HLA-A*02:02 | YGFQPTNGV | YGFRPTYGV | 868 | 281 |
| HLA-A*31:01 | GFQPTNGVGYQPYR | GFRPTYGVGHQPYR | 616 | 201 |
| HLA-A*02:06 | YGFQPTNGV | YGFRPTYGV | 174 | 61 |
| HLA-A*68:02 | YGFQPTNGV | YGFRPTYGV | 73 | 26 |
| HLA-C*12:03 | YGFQPTNGV | YGFRPTYGV | 127 | 355 |
| HLA-A*02:06 | QSYGFQPTNGV | QSYGFRPTYGV | 1121 | 425 |
| HLA-A*68:02 | QSYGFQPTNGV | QSYGFRPTYGV | 122 | 47 |
| HLA-A*02:06 | LQSYGFQPT | LQSYGFRPT | 339 | 871 |
N501Y
Reference GSTPCNGVEGFNCYFPLQSYGFQPTNGVGYQPYRVVVLSFELLHAPATVCG
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated GNRPCNGVAGPNCYSPLQSYGFRPTYGVGHQPYRVVVLSFELLHAPATVCG
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-C*03:02 | QSYGFQPTN | QSYGFRPTY | 12903 | 18 |
| HLA-B*15:01 | LQSYGFQPTN | LQSYGFRPTY | 6135 | 9 |
| HLA-C*16:01 | QSYGFQPTN | QSYGFRPTY | 10868 | 16 |
| HLA-A*29:02 | QSYGFQPTN | QSYGFRPTY | 26152 | 43 |
| HLA-B*15:03 | QSYGFQPTN | QSYGFRPTY | 6543 | 11 |
| HLA-B*15:03 | LQSYGFQPTN | LQSYGFRPTY | 2045 | 4 |
| HLA-C*12:03 | QSYGFQPTN | QSYGFRPTY | 12696 | 25 |
| HLA-B*15:03 | PLQSYGFQPTN | PLQSYGFRPTY | 22802 | 60 |
| HLA-C*14:02 | TNGVGYQPY | TYGVGHQPY | 5468 | 16 |
| HLA-C*14:02 | SYGFQPTN | SYGFRPTY | 20230 | 66 |
| HLA-C*03:02 | LQSYGFQPTN | LQSYGFRPTY | 23421 | 125 |
| HLA-B*15:01 | FQPTNGVGY | FRPTYGVGH | 166 | 29043 |
| HLA-A*30:02 | QSYGFQPTN | QSYGFRPTY | 5297 | 32 |
| HLA-B*35:01 | QSYGFQPTN | QSYGFRPTY | 24615 | 156 |
| HLA-C*03:02 | YFPLQSYGFQPTN | YSPLQSYGFRPTY | 27639 | 179 |
| HLA-C*16:01 | LQSYGFQPTN | LQSYGFRPTY | 27149 | 187 |
| HLA-A*30:02 | LQSYGFQPTN | LQSYGFRPTY | 7508 | 54 |
| HLA-B*15:01 | QSYGFQPTN | QSYGFRPTY | 21529 | 157 |
| HLA-C*12:02 | QSYGFQPTN | QSYGFRPTY | 21029 | 169 |
| HLA-B*15:03 | FPLQSYGFQPTN | SPLQSYGFRPTY | 20159 | 178 |
| HLA-B*15:03 | YFPLQSYGFQPTN | YSPLQSYGFRPTY | 22168 | 197 |
| HLA-A*30:02 | FPLQSYGFQPTN | SPLQSYGFRPTY | 21475 | 197 |
| HLA-A*29:02 | LQSYGFQPTN | LQSYGFRPTY | 28458 | 265 |
| HLA-C*16:01 | YFPLQSYGFQPTN | YSPLQSYGFRPTY | 29358 | 276 |
| HLA-B*15:03 | FQPTNGVGY | FRPTYGVGH | 63 | 6390 |
| HLA-A*29:02 | GFQPTNGVGY | GFRPTYGVGH | 188 | 18753 |
| HLA-C*14:02 | QSYGFQPTN | QSYGFRPTY | 25941 | 277 |
| HLA-C*12:03 | LQSYGFQPTN | LQSYGFRPTY | 26641 | 325 |
| HLA-B*15:03 | LQSYGFQPTNG | LQSYGFRPTYG | 6387 | 82 |
| HLA-A*32:01 | QSYGFQPTN | QSYGFRPTY | 16619 | 226 |
| HLA-A*11:01 | QSYGFQPTN | QSYGFRPTY | 22068 | 301 |
| HLA-C*02:02 | QSYGFQPTN | QSYGFRPTY | 26598 | 412 |
| HLA-C*02:10 | QSYGFQPTN | QSYGFRPTY | 26598 | 412 |
| HLA-A*30:02 | YFPLQSYGFQPTN | YSPLQSYGFRPTY | 18743 | 373 |
| HLA-A*29:02 | YFPLQSYGFQPTN | YSPLQSYGFRPTY | 19514 | 389 |
| HLA-B*15:01 | LQSYGFQPTNGVGY | LQSYGFRPTYGVGH | 180 | 8335 |
| HLA-B*15:01 | FQPTNGVGYQPY | FRPTYGVGHQPY | 290 | 11634 |
| HLA-C*03:02 | NGVGYQPY | YGVGHQPY | 10463 | 322 |
| HLA-B*15:03 | YGFQPTNGVGY | YGFRPTYGVGH | 368 | 10641 |
| HLA-A*30:02 | GFQPTNGVGY | GFRPTYGVGH | 169 | 4730 |
| HLA-A*30:02 | QSYGFQPTNGVGY | QSYGFRPTYGVGH | 258 | 7114 |
| HLA-B*15:03 | LQSYGFQPTNGVGY | LQSYGFRPTYGVGH | 113 | 2917 |
| HLA-C*16:01 | NGVGYQPY | YGVGHQPY | 8669 | 488 |
| HLA-C*07:02 | FQPTNGVGYQPY | FRPTYGVGHQPY | 6169 | 356 |
| HLA-B*57:01 | QSYGFQPTN | QSYGFRPTY | 4070 | 293 |
| HLA-A*30:01 | QSYGFQPTN | QSYGFRPTY | 3720 | 308 |
| HLA-A*02:11 | NGVGYQPYRV | YGVGHQPYRV | 5989 | 500 |
| HLA-B*15:03 | NGVGYQPY | YGVGHQPY | 4677 | 418 |
| HLA-B*57:01 | LQSYGFQPTN | LQSYGFRPTY | 3839 | 375 |
| HLA-C*14:02 | FQPTNGVGYQPY | FRPTYGVGHQPY | 3107 | 366 |
| HLA-A*29:02 | TNGVGYQPY | TYGVGHQPY | 2031 | 243 |
| HLA-A*31:01 | TNGVGYQPYR | TYGVGHQPYR | 811 | 103 |
| HLA-A*33:03 | TNGVGYQPYR | TYGVGHQPYR | 573 | 79 |
| HLA-B*15:03 | FQPTNGVGYQPY | FRPTYGVGHQPY | 183 | 1236 |
| HLA-A*31:01 | QPTNGVGYQPYR | RPTYGVGHQPYR | 3100 | 478 |
| HLA-B*58:01 | QSYGFQPTN | QSYGFRPTY | 1532 | 249 |
| HLA-A*23:01 | SYGFQPTNGV | SYGFRPTYGV | 2697 | 483 |
| HLA-A*02:11 | YGFQPTNGV | YGFRPTYGV | 307 | 59 |
| HLA-B*15:03 | LQSYGFQPTNGV | LQSYGFRPTYGV | 1971 | 442 |
| HLA-A*68:01 | PTNGVGYQPYR | PTYGVGHQPYR | 1073 | 303 |
| HLA-A*33:03 | PTNGVGYQPYR | PTYGVGHQPYR | 1345 | 417 |
| HLA-A*02:02 | YGFQPTNGV | YGFRPTYGV | 868 | 281 |
| HLA-A*31:01 | GFQPTNGVGYQPYR | GFRPTYGVGHQPYR | 616 | 201 |
| HLA-A*29:02 | PTNGVGYQPY | PTYGVGHQPY | 1211 | 401 |
| HLA-A*31:01 | PTNGVGYQPYR | PTYGVGHQPYR | 1377 | 470 |
| HLA-A*02:06 | YGFQPTNGV | YGFRPTYGV | 174 | 61 |
| HLA-A*68:01 | TNGVGYQPYR | TYGVGHQPYR | 263 | 740 |
| HLA-A*68:02 | YGFQPTNGV | YGFRPTYGV | 73 | 26 |
| HLA-C*12:03 | YGFQPTNGV | YGFRPTYGV | 127 | 355 |
| HLA-A*02:06 | QSYGFQPTNGV | QSYGFRPTYGV | 1121 | 425 |
| HLA-A*68:02 | QSYGFQPTNGV | QSYGFRPTYGV | 122 | 47 |
| HLA-A*30:02 | PTNGVGYQPY | PTYGVGHQPY | 465 | 205 |
| HLA-A*31:01 | NGVGYQPYR | YGVGHQPYR | 438 | 207 |
Y505H
Reference CNGVEGFNCYFPLQSYGFQPTNGVGYQPYRVVVLSFELLHAPATVCGPKKS
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated CNGVAGPNCYSPLQSYGFRPTYGVGHQPYRVVVLSFELLHAPATVCGPKKS
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-C*14:02 | TNGVGYQPY | TYGVGHQPY | 5468 | 16 |
| HLA-B*15:01 | FQPTNGVGY | FRPTYGVGH | 166 | 29043 |
| HLA-B*15:03 | FQPTNGVGY | FRPTYGVGH | 63 | 6390 |
| HLA-A*29:02 | GFQPTNGVGY | GFRPTYGVGH | 188 | 18753 |
| HLA-B*15:01 | LQSYGFQPTNGVGY | LQSYGFRPTYGVGH | 180 | 8335 |
| HLA-C*14:02 | GYQPYRVVV | GHQPYRVVV | 78 | 3539 |
| HLA-B*15:01 | FQPTNGVGYQPY | FRPTYGVGHQPY | 290 | 11634 |
| HLA-C*03:02 | NGVGYQPY | YGVGHQPY | 10463 | 322 |
| HLA-A*24:02 | GYQPYRVVVLSF | GHQPYRVVVLSF | 220 | 6955 |
| HLA-A*23:01 | GYQPYRVVVLSF | GHQPYRVVVLSF | 107 | 3241 |
| HLA-B*15:03 | YGFQPTNGVGY | YGFRPTYGVGH | 368 | 10641 |
| HLA-A*30:02 | GFQPTNGVGY | GFRPTYGVGH | 169 | 4730 |
| HLA-A*30:02 | QSYGFQPTNGVGY | QSYGFRPTYGVGH | 258 | 7114 |
| HLA-B*15:03 | LQSYGFQPTNGVGY | LQSYGFRPTYGVGH | 113 | 2917 |
| HLA-C*14:02 | GYQPYRVVVL | GHQPYRVVVL | 213 | 4430 |
| HLA-C*14:02 | VGYQPYRVVV | VGHQPYRVVV | 413 | 7561 |
| HLA-C*16:01 | NGVGYQPY | YGVGHQPY | 8669 | 488 |
| HLA-C*07:02 | FQPTNGVGYQPY | FRPTYGVGHQPY | 6169 | 356 |
| HLA-A*02:11 | YQPYRVVVL | HQPYRVVVL | 332 | 4009 |
| HLA-A*02:11 | NGVGYQPYRV | YGVGHQPYRV | 5989 | 500 |
| HLA-B*15:03 | NGVGYQPY | YGVGHQPY | 4677 | 418 |
| HLA-A*02:06 | YQPYRVVVL | HQPYRVVVL | 223 | 2206 |
| HLA-C*14:02 | FQPTNGVGYQPY | FRPTYGVGHQPY | 3107 | 366 |
| HLA-A*29:02 | TNGVGYQPY | TYGVGHQPY | 2031 | 243 |
| HLA-A*31:01 | TNGVGYQPYR | TYGVGHQPYR | 811 | 103 |
| HLA-A*02:02 | YQPYRVVVL | HQPYRVVVL | 360 | 2683 |
| HLA-A*33:03 | TNGVGYQPYR | TYGVGHQPYR | 573 | 79 |
| HLA-B*15:03 | FQPTNGVGYQPY | FRPTYGVGHQPY | 183 | 1236 |
| HLA-A*31:01 | QPTNGVGYQPYR | RPTYGVGHQPYR | 3100 | 478 |
| HLA-C*01:02 | YQPYRVVVL | HQPYRVVVL | 41 | 241 |
| HLA-A*02:06 | YQPYRVVVLSFEL | HQPYRVVVLSFEL | 430 | 2495 |
| HLA-C*03:03 | YQPYRVVVL | HQPYRVVVL | 410 | 2128 |
| HLA-C*03:04 | YQPYRVVVL | HQPYRVVVL | 410 | 2128 |
| HLA-C*07:02 | YQPYRVVVL | HQPYRVVVL | 135 | 590 |
| HLA-C*12:03 | YQPYRVVVL | HQPYRVVVL | 132 | 555 |
| HLA-A*68:01 | PTNGVGYQPYR | PTYGVGHQPYR | 1073 | 303 |
| HLA-C*14:02 | YQPYRVVVL | HQPYRVVVL | 102 | 347 |
| HLA-C*16:01 | YQPYRVVVL | HQPYRVVVL | 214 | 726 |
| HLA-A*33:03 | PTNGVGYQPYR | PTYGVGHQPYR | 1345 | 417 |
| HLA-A*31:01 | GFQPTNGVGYQPYR | GFRPTYGVGHQPYR | 616 | 201 |
| HLA-A*29:02 | PTNGVGYQPY | PTYGVGHQPY | 1211 | 401 |
| HLA-C*12:03 | VGYQPYRVV | VGHQPYRVV | 398 | 1196 |
| HLA-A*31:01 | PTNGVGYQPYR | PTYGVGHQPYR | 1377 | 470 |
| HLA-A*68:01 | TNGVGYQPYR | TYGVGHQPYR | 263 | 740 |
| HLA-A*23:01 | YQPYRVVVLSF | HQPYRVVVLSF | 465 | 1280 |
| HLA-A*30:02 | PTNGVGYQPY | PTYGVGHQPY | 465 | 205 |
| HLA-A*31:01 | NGVGYQPYR | YGVGHQPYR | 438 | 207 |
D614G
Reference PCSFGGVSVITPGTNTSNQVAVLYQDVNCTEVPVAIHADQLTPTWRVYSTG
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated PCSFGGVSVITPGTNTSNQVAVLYQGVNCTEVPVAIHADQLTPTWRVYSTG
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-C*05:01 | YQDVNCTEV | YQGVNCTEV | 82 | 8597 |
| HLA-C*08:02 | YQDVNCTEV | YQGVNCTEV | 89 | 8066 |
| HLA-A*02:11 | DVNCTEVPV | GVNCTEVPV | 2140 | 72 |
| HLA-A*02:02 | DVNCTEVPV | GVNCTEVPV | 4099 | 213 |
| HLA-A*68:02 | DVNCTEVPV | GVNCTEVPV | 36 | 659 |
| HLA-A*02:06 | DVNCTEVPV | GVNCTEVPV | 3807 | 208 |
| HLA-A*68:02 | QVAVLYQDV | QVAVLYQGV | 183 | 13 |
| HLA-A*68:02 | NQVAVLYQDV | NQVAVLYQGV | 650 | 52 |
| HLA-A*02:06 | QVAVLYQDV | QVAVLYQGV | 1312 | 148 |
| HLA-A*02:11 | QVAVLYQDV | QVAVLYQGV | 2169 | 268 |
| HLA-A*68:02 | NTSNQVAVLYQDV | NTSNQVAVLYQGV | 1858 | 231 |
| HLA-A*68:02 | TSNQVAVLYQDV | TSNQVAVLYQGV | 3281 | 457 |
| HLA-A*02:02 | QVAVLYQDV | QVAVLYQGV | 1107 | 188 |
| HLA-A*02:01 | YQDVNCTEV | YQGVNCTEV | 75 | 315 |
| HLA-A*02:06 | YQDVNCTEV | YQGVNCTEV | 17 | 57 |
| HLA-A*02:11 | VLYQDVNCT | VLYQGVNCT | 224 | 74 |
| HLA-A*02:11 | YQDVNCTEV | YQGVNCTEV | 31 | 93 |
| HLA-A*02:11 | VLYQDVNCTEVP | VLYQGVNCTEVP | 1000 | 402 |
| HLA-A*02:02 | NQVAVLYQDV | NQVAVLYQGV | 269 | 116 |
| HLA-A*02:11 | AVLYQDVNCTEV | AVLYQGVNCTEV | 500 | 238 |
| HLA-A*02:01 | NQVAVLYQDV | NQVAVLYQGV | 558 | 267 |
| HLA-A*02:06 | NQVAVLYQDV | NQVAVLYQGV | 36 | 18 |
H655Y
Reference TPTWRVYSTGSNVFQTRAGCLIGAEHVNNSYECDIPIGAGICASYQTQTNS
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated TPTWRVYSTGSNVFQTRAGCLIGAEYVNNSYECDIPIGAGICASYQTQTKS
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-B*15:01 | RAGCLIGAEH | RAGCLIGAEY | 7377 | 156 |
| HLA-A*30:02 | RAGCLIGAEH | RAGCLIGAEY | 1881 | 43 |
| HLA-B*15:01 | FQTRAGCLIGAEH | FQTRAGCLIGAEY | 7512 | 178 |
| HLA-B*15:03 | FQTRAGCLIGAEH | FQTRAGCLIGAEY | 5076 | 125 |
| HLA-A*30:02 | AGCLIGAEH | AGCLIGAEY | 5331 | 179 |
| HLA-B*15:03 | RAGCLIGAEH | RAGCLIGAEY | 5254 | 179 |
| HLA-A*30:02 | QTRAGCLIGAEH | QTRAGCLIGAEY | 8425 | 342 |
| HLA-B*15:03 | AGCLIGAEH | AGCLIGAEY | 9025 | 461 |
| HLA-A*02:11 | HVNNSYECDIPI | YVNNSYECDIPI | 1593 | 109 |
| HLA-A*02:01 | HVNNSYECDIPI | YVNNSYECDIPI | 2384 | 226 |
| HLA-A*02:06 | HVNNSYECDIPI | YVNNSYECDIPI | 837 | 92 |
| HLA-A*02:02 | HVNNSYECDIPI | YVNNSYECDIPI | 1466 | 187 |
| HLA-A*02:11 | CLIGAEHV | CLIGAEYV | 983 | 221 |
| HLA-B*15:03 | AEHVNNSY | AEYVNNSY | 986 | 267 |
| HLA-B*18:01 | AEHVNNSY | AEYVNNSY | 902 | 286 |
N679K
Reference EHVNNSYECDIPIGAGICASYQTQTNSPRRARSVASQSIIAYTMSLGAENS
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated EYVNNSYECDIPIGAGICASYQTQTKSHRRARSVASQSIIAYTMSLGAENS
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-A*30:01 | NSPRRARSV | KSHRRARSV | 7195 | 22 |
| HLA-A*68:01 | CASYQTQTN | CASYQTQTK | 28102 | 123 |
| HLA-B*07:02 | TNSPRRARSVA | TKSHRRARSVA | 125 | 19196 |
| HLA-B*07:02 | NSPRRARSVA | KSHRRARSVA | 69 | 5330 |
| HLA-A*11:01 | CASYQTQTN | CASYQTQTK | 32767 | 431 |
| HLA-B*07:02 | TQTNSPRRARSVA | TQTKSHRRARSVA | 270 | 17591 |
| HLA-B*07:02 | QTNSPRRARSVA | QTKSHRRARSVA | 249 | 13621 |
| HLA-B*42:01 | TNSPRRARSVA | TKSHRRARSVA | 437 | 22045 |
| HLA-B*42:01 | NSPRRARSVA | KSHRRARSVA | 217 | 9050 |
| HLA-A*30:01 | NSPRRARSVAS | KSHRRARSVAS | 4546 | 185 |
| HLA-A*30:01 | NSPRRARSVA | KSHRRARSVA | 630 | 27 |
| HLA-C*15:02 | NSPRRARSV | KSHRRARSV | 4094 | 215 |
| HLA-A*68:01 | QTNSPRRAR | QTKSHRRAR | 95 | 1735 |
| HLA-C*14:02 | SYQTQTNSP | SYQTQTKSH | 3691 | 214 |
| HLA-A*68:01 | TQTNSPRRAR | TQTKSHRRAR | 254 | 4244 |
| HLA-A*68:01 | CASYQTQTNSPR | CASYQTQTKSHR | 142 | 713 |
| HLA-C*16:01 | NSPRRARSV | KSHRRARSV | 1247 | 297 |
| HLA-A*11:01 | ASYQTQTNSPR | ASYQTQTKSHR | 346 | 1426 |
| HLA-A*31:01 | QTQTNSPRR | QTQTKSHRR | 355 | 96 |
| HLA-A*33:03 | YQTQTNSPRR | YQTQTKSHRR | 967 | 273 |
| HLA-A*33:03 | YQTQTNSPRRAR | YQTQTKSHRRAR | 306 | 1077 |
| HLA-A*31:01 | YQTQTNSPRR | YQTQTKSHRR | 965 | 276 |
| HLA-A*33:03 | TQTNSPRRAR | TQTKSHRRAR | 195 | 659 |
| HLA-A*33:03 | QTQTNSPRR | QTQTKSHRR | 445 | 143 |
| HLA-A*31:01 | ASYQTQTNSPRR | ASYQTQTKSHRR | 433 | 144 |
| HLA-A*33:03 | QTNSPRRAR | QTKSHRRAR | 48 | 144 |
| HLA-A*30:01 | QTNSPRRAR | QTKSHRRAR | 1146 | 463 |
| HLA-A*31:01 | YQTQTNSPRRAR | YQTQTKSHRRAR | 317 | 771 |
| HLA-A*31:01 | SYQTQTNSPR | SYQTQTKSHR | 62 | 147 |
| HLA-A*33:03 | SYQTQTNSPR | SYQTQTKSHR | 130 | 304 |
| HLA-A*31:01 | TQTNSPRRAR | TQTKSHRRAR | 144 | 336 |
| HLA-A*31:01 | QTNSPRRAR | QTKSHRRAR | 34 | 79 |
| HLA-A*31:01 | ASYQTQTNSPR | ASYQTQTKSHR | 139 | 315 |
| HLA-A*31:01 | YQTQTNSPR | YQTQTKSHR | 441 | 982 |
| HLA-A*31:01 | SYQTQTNSPRRAR | SYQTQTKSHRRAR | 193 | 427 |
| HLA-A*33:03 | SYQTQTNSPRRAR | SYQTQTKSHRRAR | 448 | 972 |
P681H
Reference VNNSYECDIPIGAGICASYQTQTNSPRRARSVASQSIIAYTMSLGAENSVA
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated VNNSYECDIPIGAGICASYQTQTKSHRRARSVASQSIIAYTMSLGAENSVA
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-B*07:02 | SPRRARSVA | SHRRARSVA | 4 | 4721 |
| HLA-B*42:01 | SPRRARSVA | SHRRARSVA | 8 | 6169 |
| HLA-B*07:02 | SPRRARSVASQSI | SHRRARSVASQSI | 17 | 11433 |
| HLA-B*07:02 | SPRRARSV | SHRRARSV | 24 | 12728 |
| HLA-B*07:02 | SPRRARSVAS | SHRRARSVAS | 37 | 14758 |
| HLA-A*30:01 | NSPRRARSV | KSHRRARSV | 7195 | 22 |
| HLA-B*07:02 | SPRRARSVASQSII | SHRRARSVASQSII | 55 | 17736 |
| HLA-B*42:01 | SPRRARSV | SHRRARSV | 38 | 11622 |
| HLA-B*42:01 | SPRRARSVASQSI | SHRRARSVASQSI | 48 | 14418 |
| HLA-B*07:02 | TNSPRRARSVA | TKSHRRARSVA | 125 | 19196 |
| HLA-B*42:01 | SPRRARSVAS | SHRRARSVAS | 158 | 17461 |
| HLA-B*07:02 | NSPRRARSVA | KSHRRARSVA | 69 | 5330 |
| HLA-B*42:01 | SPRRARSVASQSII | SHRRARSVASQSII | 321 | 21521 |
| HLA-B*07:02 | TQTNSPRRARSVA | TQTKSHRRARSVA | 270 | 17591 |
| HLA-B*07:02 | QTNSPRRARSVA | QTKSHRRARSVA | 249 | 13621 |
| HLA-B*42:01 | TNSPRRARSVA | TKSHRRARSVA | 437 | 22045 |
| HLA-B*42:01 | NSPRRARSVA | KSHRRARSVA | 217 | 9050 |
| HLA-A*30:01 | NSPRRARSVAS | KSHRRARSVAS | 4546 | 185 |
| HLA-A*30:01 | NSPRRARSVA | KSHRRARSVA | 630 | 27 |
| HLA-C*15:02 | NSPRRARSV | KSHRRARSV | 4094 | 215 |
| HLA-A*68:01 | QTNSPRRAR | QTKSHRRAR | 95 | 1735 |
| HLA-C*14:02 | SYQTQTNSP | SYQTQTKSH | 3691 | 214 |
| HLA-A*68:01 | TQTNSPRRAR | TQTKSHRRAR | 254 | 4244 |
| HLA-B*08:01 | SPRRARSVA | SHRRARSVA | 372 | 3831 |
| HLA-B*08:01 | SPRRARSV | SHRRARSV | 428 | 2448 |
| HLA-A*68:01 | CASYQTQTNSPR | CASYQTQTKSHR | 142 | 713 |
| HLA-C*16:01 | NSPRRARSV | KSHRRARSV | 1247 | 297 |
| HLA-A*11:01 | ASYQTQTNSPR | ASYQTQTKSHR | 346 | 1426 |
| HLA-A*31:01 | QTQTNSPRR | QTQTKSHRR | 355 | 96 |
| HLA-A*33:03 | YQTQTNSPRR | YQTQTKSHRR | 967 | 273 |
| HLA-A*33:03 | YQTQTNSPRRAR | YQTQTKSHRRAR | 306 | 1077 |
| HLA-A*31:01 | YQTQTNSPRR | YQTQTKSHRR | 965 | 276 |
| HLA-A*33:03 | TQTNSPRRAR | TQTKSHRRAR | 195 | 659 |
| HLA-A*33:03 | QTQTNSPRR | QTQTKSHRR | 445 | 143 |
| HLA-A*31:01 | ASYQTQTNSPRR | ASYQTQTKSHRR | 433 | 144 |
| HLA-A*33:03 | QTNSPRRAR | QTKSHRRAR | 48 | 144 |
| HLA-A*30:01 | QTNSPRRAR | QTKSHRRAR | 1146 | 463 |
| HLA-A*31:01 | YQTQTNSPRRAR | YQTQTKSHRRAR | 317 | 771 |
| HLA-A*31:01 | SYQTQTNSPR | SYQTQTKSHR | 62 | 147 |
| HLA-A*33:03 | SYQTQTNSPR | SYQTQTKSHR | 130 | 304 |
| HLA-A*31:01 | TQTNSPRRAR | TQTKSHRRAR | 144 | 336 |
| HLA-A*31:01 | QTNSPRRAR | QTKSHRRAR | 34 | 79 |
| HLA-A*31:01 | ASYQTQTNSPR | ASYQTQTKSHR | 139 | 315 |
| HLA-A*31:01 | YQTQTNSPR | YQTQTKSHR | 441 | 982 |
| HLA-A*31:01 | SYQTQTNSPRRAR | SYQTQTKSHRRAR | 193 | 427 |
| HLA-A*33:03 | SYQTQTNSPRRAR | SYQTQTKSHRRAR | 448 | 972 |
N764K
Reference TMYICGDSTECSNLLLQYGSFCTQLNRALTGIAVEQDKNTQEVFAQVKQIY
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated TMYICGDSTECSNLLLQYGSFCTQLKRALTGIAVEQDKNTQEVFAQVKQIY
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-A*03:02 | LLQYGSFCTQLN | LLQYGSFCTQLK | 29153 | 102 |
| HLA-A*11:01 | LQYGSFCTQLN | LQYGSFCTQLK | 32767 | 191 |
| HLA-A*03:01 | LQYGSFCTQLN | LQYGSFCTQLK | 28204 | 214 |
| HLA-A*68:01 | YGSFCTQLN | YGSFCTQLK | 23773 | 187 |
| HLA-A*03:02 | LQYGSFCTQLN | LQYGSFCTQLK | 31227 | 331 |
| HLA-A*03:02 | LLLQYGSFCTQLN | LLLQYGSFCTQLK | 32767 | 377 |
| HLA-A*11:01 | YGSFCTQLN | YGSFCTQLK | 32767 | 386 |
| HLA-A*03:01 | LLQYGSFCTQLN | LLQYGSFCTQLK | 29424 | 350 |
| HLA-A*03:01 | LLLQYGSFCTQLN | LLLQYGSFCTQLK | 32355 | 410 |
| HLA-A*02:11 | QLNRALTGI | QLKRALTGI | 27 | 440 |
| HLA-A*02:11 | FCTQLNRALTGI | FCTQLKRALTGI | 63 | 898 |
| HLA-B*08:01 | CTQLNRAL | CTQLKRAL | 3325 | 279 |
| HLA-A*02:01 | FCTQLNRALTGI | FCTQLKRALTGI | 493 | 4878 |
| HLA-A*02:02 | FCTQLNRALTGI | FCTQLKRALTGI | 62 | 503 |
| HLA-A*02:02 | QLNRALTGI | QLKRALTGI | 45 | 307 |
| HLA-C*03:03 | FCTQLNRAL | FCTQLKRAL | 138 | 752 |
| HLA-C*03:04 | FCTQLNRAL | FCTQLKRAL | 138 | 752 |
| HLA-A*02:02 | TQLNRALTGI | TQLKRALTGI | 180 | 882 |
| HLA-A*02:11 | TQLNRALTGI | TQLKRALTGI | 267 | 1284 |
| HLA-B*27:05 | NRALTGIAV | KRALTGIAV | 1251 | 276 |
| HLA-A*02:11 | QLNRALTGIAV | QLKRALTGIAV | 259 | 1123 |
| HLA-C*03:02 | FCTQLNRAL | FCTQLKRAL | 361 | 1438 |
| HLA-C*14:02 | SFCTQLNRAL | SFCTQLKRAL | 234 | 511 |
| HLA-A*02:06 | TQLNRALTGI | TQLKRALTGI | 54 | 113 |
D796Y
Reference AVEQDKNTQEVFAQVKQIYKTPPIKDFGGFNFSQILPDPSKPSKRSFIEDL
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated AVEQDKNTQEVFAQVKQIYKTPPIKYFGGFNFSQILPDPSKPSKRSFIEDL
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-A*23:01 | KDFGGFNF | KYFGGFNF | 15419 | 23 |
| HLA-A*23:01 | KDFGGFNFSQI | KYFGGFNFSQI | 15932 | 37 |
| HLA-A*24:02 | KDFGGFNF | KYFGGFNF | 21008 | 55 |
| HLA-C*14:02 | IYKTPPIKD | IYKTPPIKY | 16538 | 55 |
| HLA-A*24:02 | KDFGGFNFSQI | KYFGGFNFSQI | 20251 | 68 |
| HLA-B*15:03 | KQIYKTPPIKD | KQIYKTPPIKY | 7773 | 29 |
| HLA-A*23:01 | KDFGGFNFSQIL | KYFGGFNFSQIL | 15831 | 63 |
| HLA-C*14:02 | KDFGGFNF | KYFGGFNF | 22081 | 90 |
| HLA-A*24:02 | KDFGGFNFSQIL | KYFGGFNFSQIL | 21565 | 126 |
| HLA-B*15:01 | KQIYKTPPIKD | KQIYKTPPIKY | 22193 | 141 |
| HLA-C*14:02 | KDFGGFNFSQIL | KYFGGFNFSQIL | 14873 | 165 |
| HLA-A*30:01 | KDFGGFNFS | KYFGGFNFS | 9496 | 156 |
| HLA-C*14:02 | DFGGFNFSQIL | YFGGFNFSQIL | 6960 | 139 |
| HLA-A*30:02 | KQIYKTPPIKD | KQIYKTPPIKY | 15917 | 386 |
| HLA-B*15:03 | VKQIYKTPPIKD | VKQIYKTPPIKY | 16977 | 446 |
| HLA-B*15:03 | IKDFGGFNF | IKYFGGFNF | 97 | 4 |
| HLA-B*15:03 | PIKDFGGFNF | PIKYFGGFNF | 7546 | 321 |
| HLA-B*15:03 | TPPIKDFGGFNF | TPPIKYFGGFNF | 10071 | 484 |
| HLA-B*15:03 | KTPPIKDFGGFNF | KTPPIKYFGGFNF | 6427 | 331 |
| HLA-A*32:01 | KTPPIKDFGGFNF | KTPPIKYFGGFNF | 6031 | 381 |
| HLA-A*23:01 | DFGGFNFSQI | YFGGFNFSQI | 5424 | 461 |
| HLA-B*15:03 | YKTPPIKDFGGFNF | YKTPPIKYFGGFNF | 2588 | 394 |
| HLA-C*06:02 | YKTPPIKDF | YKTPPIKYF | 2488 | 460 |
| HLA-C*07:02 | YKTPPIKDF | YKTPPIKYF | 1804 | 378 |
| HLA-A*23:01 | IYKTPPIKDF | IYKTPPIKYF | 273 | 98 |
| HLA-A*24:02 | IYKTPPIKDF | IYKTPPIKYF | 375 | 147 |
| HLA-C*14:02 | IYKTPPIKDF | IYKTPPIKYF | 341 | 158 |
| HLA-B*15:03 | YKTPPIKDF | YKTPPIKYF | 92 | 43 |
Q954H
Reference SAIGKIQDSLSSTASALGKLQDVVNQNAQALNTLVKQLSSNFGAISSVLND
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated SAIGKIQDSLSSTASALGKLQDVVNHNAQALNTLVKQLSSKFGAISSVLND
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-B*38:01 | NQNAQALNTL | NHNAQALNTL | 2805 | 97 |
| HLA-A*68:02 | DVVNQNAQA | DVVNHNAQA | 703 | 344 |
N969K
Reference ALGKLQDVVNQNAQALNTLVKQLSSNFGAISSVLNDILSRLDKVEAEVQID
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated ALGKLQDVVNHNAQALNTLVKQLSSKFGAISSVLNDILSRLDKVEAEVQID
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-A*03:01 | TLVKQLSSN | TLVKQLSSK | 29207 | 56 |
| HLA-A*68:01 | NTLVKQLSSN | NTLVKQLSSK | 28063 | 86 |
| HLA-A*11:01 | TLVKQLSSN | TLVKQLSSK | 32767 | 125 |
| HLA-A*03:02 | TLVKQLSSN | TLVKQLSSK | 31915 | 131 |
| HLA-A*11:01 | NTLVKQLSSN | NTLVKQLSSK | 32767 | 147 |
| HLA-A*03:01 | ALNTLVKQLSSN | ALNTLVKQLSSK | 32767 | 154 |
| HLA-A*03:02 | ALNTLVKQLSSN | ALNTLVKQLSSK | 32767 | 304 |
| HLA-A*03:01 | NTLVKQLSSN | NTLVKQLSSK | 32767 | 482 |
| HLA-A*68:02 | SNFGAISSV | SKFGAISSV | 117 | 5329 |
| HLA-B*15:03 | SNFGAISSVL | SKFGAISSVL | 688 | 25 |
| HLA-B*08:01 | QLSSNFGAI | QLSSKFGAI | 4755 | 265 |
| HLA-C*15:02 | SNFGAISSV | SKFGAISSV | 374 | 5700 |
| HLA-A*30:01 | SSNFGAISS | SSKFGAISS | 1320 | 88 |
| HLA-B*15:03 | SNFGAISSV | SKFGAISSV | 727 | 49 |
| HLA-A*30:01 | SSNFGAISSV | SSKFGAISSV | 587 | 71 |
| HLA-A*02:06 | KQLSSNFGA | KQLSSKFGA | 62 | 269 |
| HLA-A*02:01 | KQLSSNFGAI | KQLSSKFGAI | 407 | 1567 |
| HLA-A*02:11 | KQLSSNFGA | KQLSSKFGA | 228 | 807 |
| HLA-A*02:06 | SNFGAISSV | SKFGAISSV | 361 | 1100 |
| HLA-C*06:02 | SNFGAISSV | SKFGAISSV | 1423 | 485 |
| HLA-A*02:11 | KQLSSNFGAI | KQLSSKFGAI | 152 | 428 |
| HLA-A*02:06 | KQLSSNFGAI | KQLSSKFGAI | 58 | 159 |
| HLA-A*02:02 | KQLSSNFGAI | KQLSSKFGAI | 261 | 668 |
| HLA-B*15:03 | KQLSSNFGA | KQLSSKFGA | 497 | 1076 |
N protein
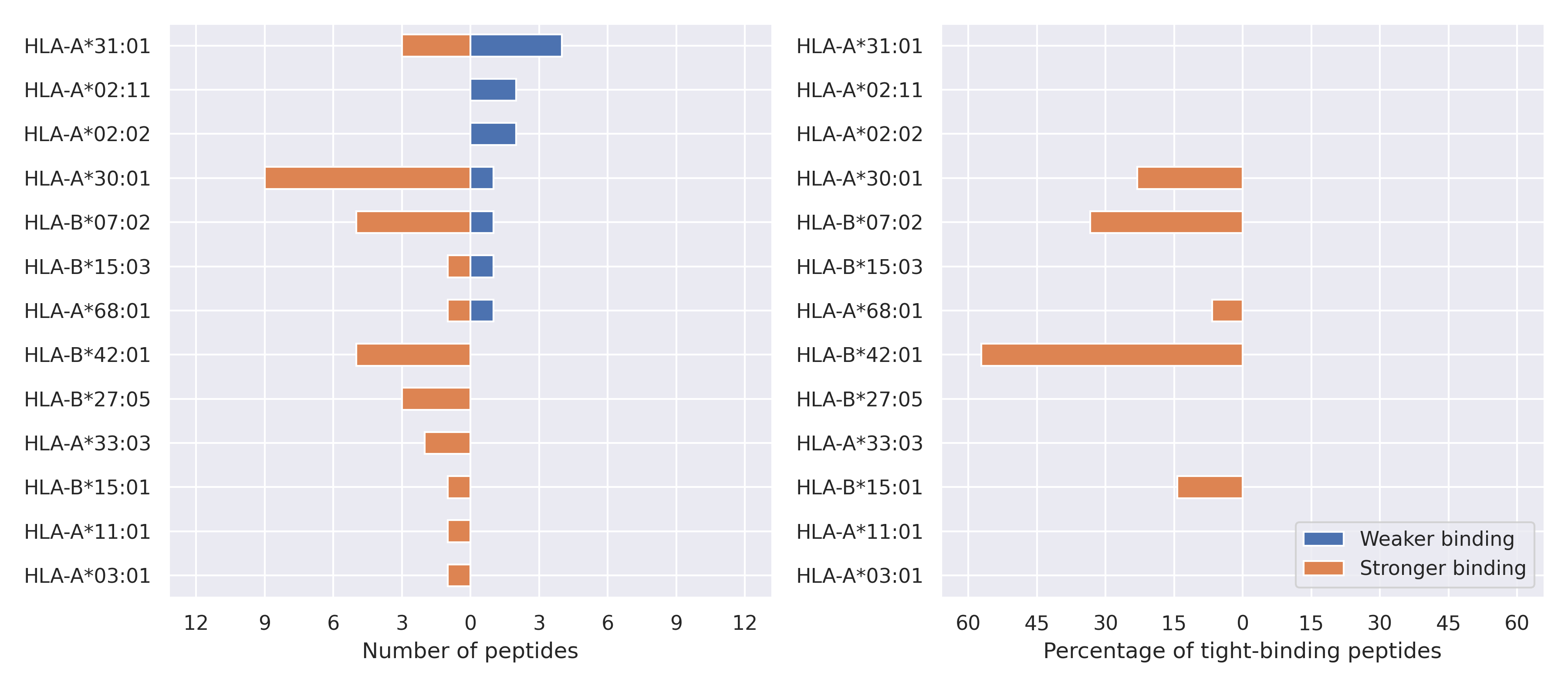
P13L
Reference MSDNGPQNQRNAPRITFGGPSDSTGSNQNGERSGARSK
|||||||||||| |||||||||||||||||||||||||
Mutated MSDNGPQNQRNALRITFGGPSDSTGSNQNGERGGARSK
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-B*42:01 | MSDNGPQNQRNAP | MSDNGPQNQRNAL | 11146 | 27 |
| HLA-B*07:02 | GPQNQRNAP | GPQNQRNAL | 2464 | 6 |
| HLA-B*42:01 | GPQNQRNAP | GPQNQRNAL | 3035 | 8 |
| HLA-B*07:02 | SDNGPQNQRNAP | SDNGPQNQRNAL | 11353 | 32 |
| HLA-B*42:01 | SDNGPQNQRNAP | SDNGPQNQRNAL | 13042 | 39 |
| HLA-B*42:01 | NGPQNQRNAP | NGPQNQRNAL | 12875 | 41 |
| HLA-B*07:02 | MSDNGPQNQRNAP | MSDNGPQNQRNAL | 13138 | 58 |
| HLA-B*07:02 | NGPQNQRNAP | NGPQNQRNAL | 14898 | 72 |
| HLA-B*07:02 | APRITFGGPS | ALRITFGGPS | 191 | 17534 |
| HLA-B*42:01 | PQNQRNAP | PQNQRNAL | 30142 | 436 |
| HLA-B*07:02 | GPQNQRNAPR | GPQNQRNALR | 13964 | 345 |
| HLA-B*27:05 | QRNAPRITF | QRNALRITF | 509 | 83 |
| HLA-B*27:05 | NQRNAPRITF | NQRNALRITF | 1716 | 350 |
| HLA-B*27:05 | GPQNQRNAPRITF | GPQNQRNALRITF | 2076 | 442 |
| HLA-B*15:01 | NQRNAPRITF | NQRNALRITF | 96 | 40 |
S33G
Reference NQRNAPRITFGGPSDSTGSNQNGERSGARSKQRRPQGLPNNTASWFTALTQ
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated NQRNALRITFGGPSDSTGSNQNGERGGARSKQRRPQGLPNNTASWFTALTQ
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-A*31:01 | RSGARSKQR | RGGARSKQR | 77 | 488 |
| HLA-A*31:01 | RSGARSKQRR | RGGARSKQRR | 254 | 1585 |
203-204 RG->KR
Reference GGSQASSRSSSRSRNSSRNSTPGSSRGTSPARMAGNGGDAALALLLLDRLNQ
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated GGSQASSRSSSRSRNSSRNSTPGSSKRTSPARMAGNGGDAALALLLLDRLNQ
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-A*68:01 | NSTPGSSRG | NSTPGSSKR | 32767 | 34 |
| HLA-A*33:03 | NSTPGSSRG | NSTPGSSKR | 32767 | 300 |
| HLA-B*15:03 | SRGTSPARM | SKRTSPARM | 2669 | 106 |
| HLA-A*31:01 | RNSTPGSSRG | RNSTPGSSKR | 7013 | 340 |
| HLA-A*30:01 | SSRNSTPGSSR | SSRNSTPGSSK | 356 | 20 |
| HLA-A*30:01 | GTSPARMAG | RTSPARMAG | 493 | 30 |
| HLA-A*31:01 | SSRNSTPGSSRG | SSRNSTPGSSKR | 3757 | 231 |
| HLA-A*30:01 | GTSPARMAGNGG | RTSPARMAGNGG | 6051 | 411 |
| HLA-A*31:01 | SSRNSTPGSSR | SSRNSTPGSSK | 127 | 1853 |
| HLA-A*30:01 | NSSRNSTPGSSR | NSSRNSTPGSSK | 4255 | 353 |
| HLA-A*30:01 | RNSTPGSSR | RNSTPGSSK | 2120 | 188 |
| HLA-A*31:01 | RNSTPGSSR | RNSTPGSSK | 123 | 1335 |
| HLA-A*03:01 | SSRNSTPGSSR | SSRNSTPGSSK | 4753 | 442 |
| HLA-A*11:01 | SSRNSTPGSSR | SSRNSTPGSSK | 2231 | 263 |
| HLA-A*68:01 | NSSRNSTPGSSR | NSSRNSTPGSSK | 270 | 1385 |
| HLA-A*30:01 | GSSRGTSPA | GSSKRTSPA | 555 | 145 |
| HLA-A*30:01 | SSRNSTPGSSRG | SSRNSTPGSSKR | 1625 | 469 |
| HLA-A*30:01 | SSRGTSPARMAG | SSKRTSPARMAG | 449 | 1031 |
S413R
Reference KQQTVTLLPAADLDDFSKQLQQSMSSADSTQA
||||||||||||||||||||||||| ||||||
Mutated KQQTVTLLPAADLDDFSKQLQQSMSRADSTQA
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-A*31:01 | KQLQQSMSS | KQLQQSMSR | 20016 | 55 |
| HLA-A*30:01 | MSSADSTQA | MSRADSTQA | 2953 | 37 |
| HLA-A*33:03 | DFSKQLQQSMSS | DFSKQLQQSMSR | 32767 | 494 |
| HLA-B*15:03 | KQLQQSMSS | KQLQQSMSR | 236 | 5105 |
| HLA-A*30:01 | SMSSADSTQA | SMSRADSTQA | 6453 | 416 |
| HLA-A*02:02 | FSKQLQQSMSSA | FSKQLQQSMSRA | 345 | 1138 |
| HLA-A*02:11 | FSKQLQQSMSSA | FSKQLQQSMSRA | 353 | 1118 |
| HLA-A*02:11 | QLQQSMSSA | QLQQSMSRA | 436 | 1175 |
| HLA-A*02:02 | SMSSADSTQA | SMSRADSTQA | 355 | 725 |
M protein
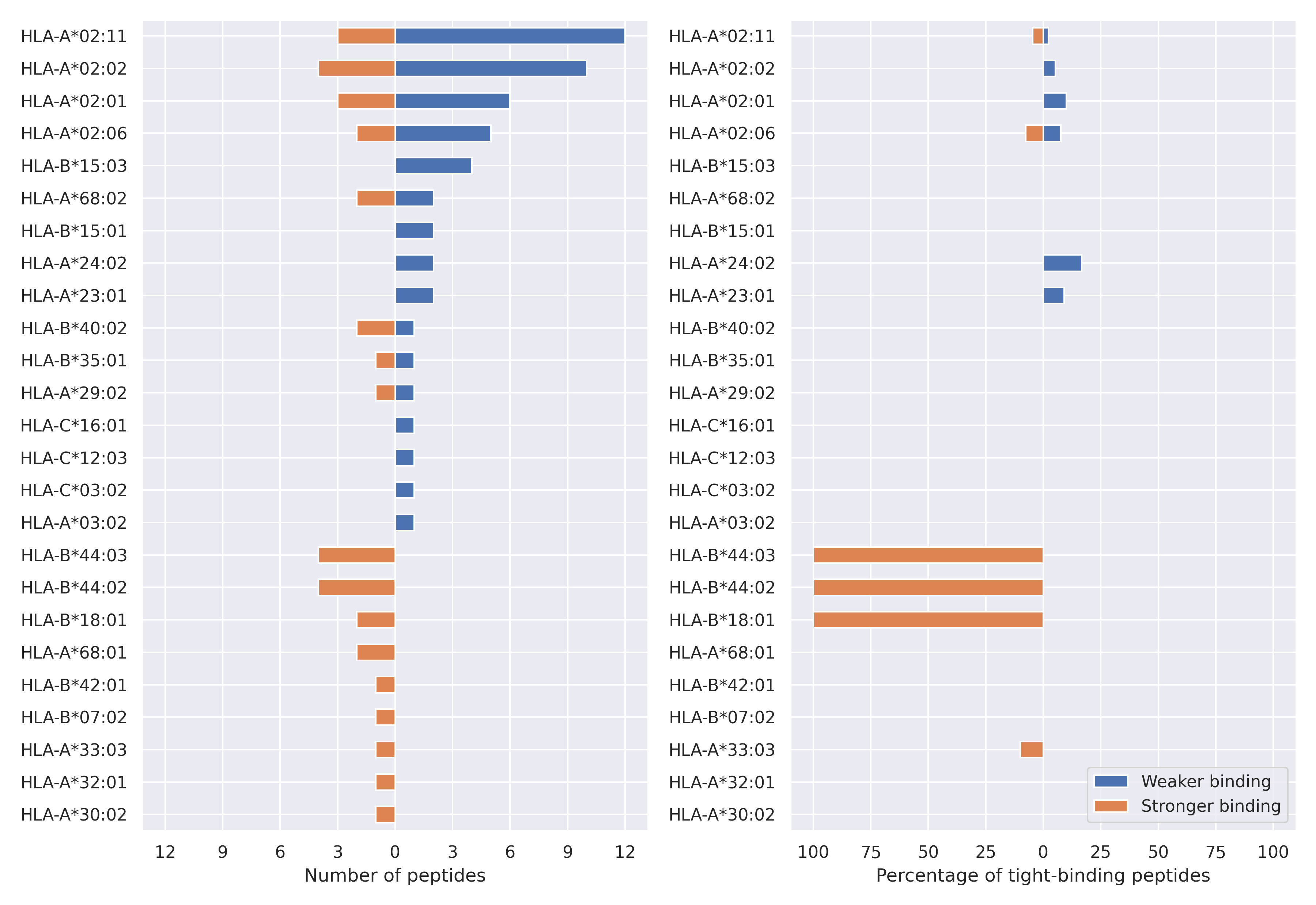
Q19E
Reference MADSNGTITVEELKKLLEQWNLVIGFLFLTWICLLQFAYANRNR
|||||||||||||||||| |||||||||||||||||||||||||
Mutated MADSNGTITVEELKKLLEEWNLVIGFLFLTWICLLQFAYANRNR
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-B*44:03 | EQWNLVIGF | EEWNLVIGF | 1268 | 15 |
| HLA-B*44:02 | EQWNLVIGF | EEWNLVIGF | 2396 | 34 |
| HLA-B*44:03 | EQWNLVIGFLF | EEWNLVIGFLF | 5200 | 85 |
| HLA-B*44:03 | EQWNLVIGFL | EEWNLVIGFL | 8038 | 135 |
| HLA-B*44:03 | EQWNLVIGFLFLTW | EEWNLVIGFLFLTW | 5966 | 126 |
| HLA-B*44:02 | EQWNLVIGFLF | EEWNLVIGFLF | 6607 | 162 |
| HLA-B*44:02 | EQWNLVIGFL | EEWNLVIGFL | 9054 | 237 |
| HLA-B*44:02 | EQWNLVIGFLFLTW | EEWNLVIGFLFLTW | 5922 | 170 |
| HLA-B*15:01 | LEQWNLVIGF | LEEWNLVIGF | 256 | 8100 |
| HLA-B*18:01 | EQWNLVIGF | EEWNLVIGF | 482 | 16 |
| HLA-A*02:06 | EQWNLVIGFL | EEWNLVIGFL | 418 | 10916 |
| HLA-B*15:03 | EQWNLVIGF | EEWNLVIGF | 101 | 2397 |
| HLA-B*18:01 | EQWNLVIGFLF | EEWNLVIGFLF | 3471 | 190 |
| HLA-B*40:02 | EQWNLVIGFL | EEWNLVIGFL | 3775 | 245 |
| HLA-B*15:03 | LEQWNLVIGF | LEEWNLVIGF | 88 | 1227 |
| HLA-B*40:02 | EQWNLVIGF | EEWNLVIGF | 1765 | 242 |
| HLA-B*15:03 | KLLEQWNLVIGF | KLLEEWNLVIGF | 365 | 2285 |
| HLA-A*02:11 | LLEQWNLVI | LLEEWNLVI | 142 | 24 |
| HLA-A*02:11 | LLEQWNLVIGFL | LLEEWNLVIGFL | 1120 | 210 |
| HLA-A*02:01 | LLEQWNLVI | LLEEWNLVI | 644 | 131 |
| HLA-A*29:02 | QWNLVIGFLF | EWNLVIGFLF | 159 | 780 |
| HLA-A*24:02 | QWNLVIGFLF | EWNLVIGFLF | 53 | 246 |
| HLA-A*23:01 | QWNLVIGFLF | EWNLVIGFLF | 18 | 79 |
| HLA-A*02:02 | LLEQWNLVI | LLEEWNLVI | 781 | 187 |
| HLA-A*02:06 | LLEQWNLVI | LLEEWNLVI | 1869 | 487 |
| HLA-B*40:02 | LEQWNLVI | LEEWNLVI | 367 | 1301 |
| HLA-A*02:02 | LLEQWNLVIGFL | LLEEWNLVIGFL | 1047 | 329 |
A63T
Reference AYANRNRFLYIIKLIFLWLLWPVTLACFVLAAVYRINWITGGIAIAMACLV
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated AYANRNRFLYIIKLIFLWLLWPVTLTCFVLAAVYRINWITGGIAIAMACLV
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-B*35:01 | LACFVLAAVY | LTCFVLAAVY | 121 | 1943 |
| HLA-A*68:01 | ACFVLAAVYR | TCFVLAAVYR | 694 | 80 |
| HLA-B*35:01 | ACFVLAAVY | TCFVLAAVY | 2289 | 331 |
| HLA-C*03:02 | LACFVLAAVY | LTCFVLAAVY | 145 | 935 |
| HLA-C*16:01 | LACFVLAAV | LTCFVLAAV | 346 | 1994 |
| HLA-A*02:01 | WLLWPVTLA | WLLWPVTLT | 33 | 190 |
| HLA-A*02:02 | WLLWPVTLA | WLLWPVTLT | 32 | 180 |
| HLA-A*02:11 | LWLLWPVTLA | LWLLWPVTLT | 66 | 363 |
| HLA-A*02:11 | IFLWLLWPVTLA | IFLWLLWPVTLT | 190 | 1034 |
| HLA-A*02:11 | LLWPVTLA | LLWPVTLT | 242 | 1313 |
| HLA-A*33:03 | ACFVLAAVYR | TCFVLAAVYR | 252 | 47 |
| HLA-A*02:02 | LWLLWPVTLA | LWLLWPVTLT | 304 | 1623 |
| HLA-C*12:03 | LACFVLAAV | LTCFVLAAV | 214 | 1116 |
| HLA-A*02:06 | WLLWPVTLA | WLLWPVTLT | 60 | 302 |
| HLA-A*02:01 | LWLLWPVTLA | LWLLWPVTLT | 228 | 1142 |
| HLA-A*02:02 | TLACFVLAAVY | TLTCFVLAAVY | 337 | 1467 |
| HLA-A*02:02 | VTLACFVLAAV | VTLTCFVLAAV | 160 | 678 |
| HLA-A*68:02 | LACFVLAAV | LTCFVLAAV | 44 | 11 |
| HLA-A*02:02 | PVTLACFVLAAV | PVTLTCFVLAAV | 479 | 1907 |
| HLA-A*02:11 | LIFLWLLWPVTLA | LIFLWLLWPVTLT | 406 | 1602 |
| HLA-A*02:02 | TLACFVLAAVYRI | TLTCFVLAAVYRI | 293 | 1148 |
| HLA-A*02:11 | LACFVLAAV | LTCFVLAAV | 122 | 32 |
| HLA-A*02:11 | WLLWPVTLA | WLLWPVTLT | 6 | 22 |
| HLA-B*15:03 | LACFVLAAVY | LTCFVLAAVY | 81 | 284 |
| HLA-A*68:02 | TLACFVLAA | TLTCFVLAA | 395 | 1376 |
| HLA-A*02:02 | FLWLLWPVTLACFV | FLWLLWPVTLTCFV | 485 | 146 |
| HLA-A*68:01 | LACFVLAAVY | LTCFVLAAVY | 1184 | 359 |
| HLA-A*02:02 | TLACFVLAA | TLTCFVLAA | 13 | 41 |
| HLA-A*02:11 | TLACFVLAAVY | TLTCFVLAAVY | 310 | 929 |
| HLA-A*02:02 | FLWLLWPVTLA | FLWLLWPVTLT | 291 | 867 |
| HLA-A*02:11 | KLIFLWLLWPVTLA | KLIFLWLLWPVTLT | 290 | 859 |
| HLA-A*29:02 | LACFVLAAVY | LTCFVLAAVY | 232 | 81 |
| HLA-A*02:02 | VTLACFVLAA | VTLTCFVLAA | 100 | 285 |
| HLA-A*02:06 | TLACFVLAAV | TLTCFVLAAV | 16 | 45 |
| HLA-A*68:02 | VTLACFVLAA | VTLTCFVLAA | 481 | 1321 |
| HLA-B*15:01 | TLACFVLAAVY | TLTCFVLAAVY | 414 | 1136 |
| HLA-A*23:01 | LWPVTLACF | LWPVTLTCF | 57 | 155 |
| HLA-A*24:02 | LWPVTLACF | LWPVTLTCF | 49 | 133 |
| HLA-A*02:11 | FLWLLWPVTLA | FLWLLWPVTLT | 25 | 67 |
| HLA-A*02:06 | TLACFVLAA | TLTCFVLAA | 40 | 107 |
| HLA-A*02:01 | LACFVLAAV | LTCFVLAAV | 471 | 179 |
| HLA-A*02:02 | TLACFVLAAV | TLTCFVLAAV | 5 | 13 |
| HLA-A*02:11 | VTLACFVLAA | VTLTCFVLAA | 97 | 245 |
| HLA-A*02:01 | FLWLLWPVTLA | FLWLLWPVTLT | 107 | 269 |
| HLA-A*02:06 | FLWLLWPVTLA | FLWLLWPVTLT | 495 | 1240 |
| HLA-A*02:11 | TLACFVLAA | TLTCFVLAA | 8 | 20 |
| HLA-A*02:11 | VTLACFVLAAV | VTLTCFVLAAV | 128 | 318 |
| HLA-A*02:01 | TLACFVLAAV | TLTCFVLAAV | 11 | 27 |
| HLA-A*02:02 | LACFVLAAV | LTCFVLAAV | 198 | 81 |
| HLA-A*02:01 | TLACFVLAA | TLTCFVLAA | 25 | 61 |
| HLA-A*02:01 | VTLACFVLAAV | VTLTCFVLAAV | 436 | 1029 |
| HLA-B*07:02 | WPVTLACFVL | WPVTLTCFVL | 362 | 155 |
| HLA-A*02:01 | FLWLLWPVTLACFV | FLWLLWPVTLTCFV | 329 | 142 |
| HLA-A*02:11 | TLACFVLAAV | TLTCFVLAAV | 4 | 9 |
| HLA-A*30:02 | LACFVLAAVY | LTCFVLAAVY | 218 | 98 |
| HLA-A*02:06 | LACFVLAAV | LTCFVLAAV | 51 | 23 |
| HLA-A*03:02 | TLACFVLAAVYR | TLTCFVLAAVYR | 480 | 1047 |
| HLA-A*68:02 | TLACFVLAAV | TLTCFVLAAV | 26 | 12 |
| HLA-B*42:01 | WPVTLACFV | WPVTLTCFV | 209 | 102 |
| HLA-A*32:01 | LLWPVTLACF | LLWPVTLTCF | 427 | 212 |
E protein
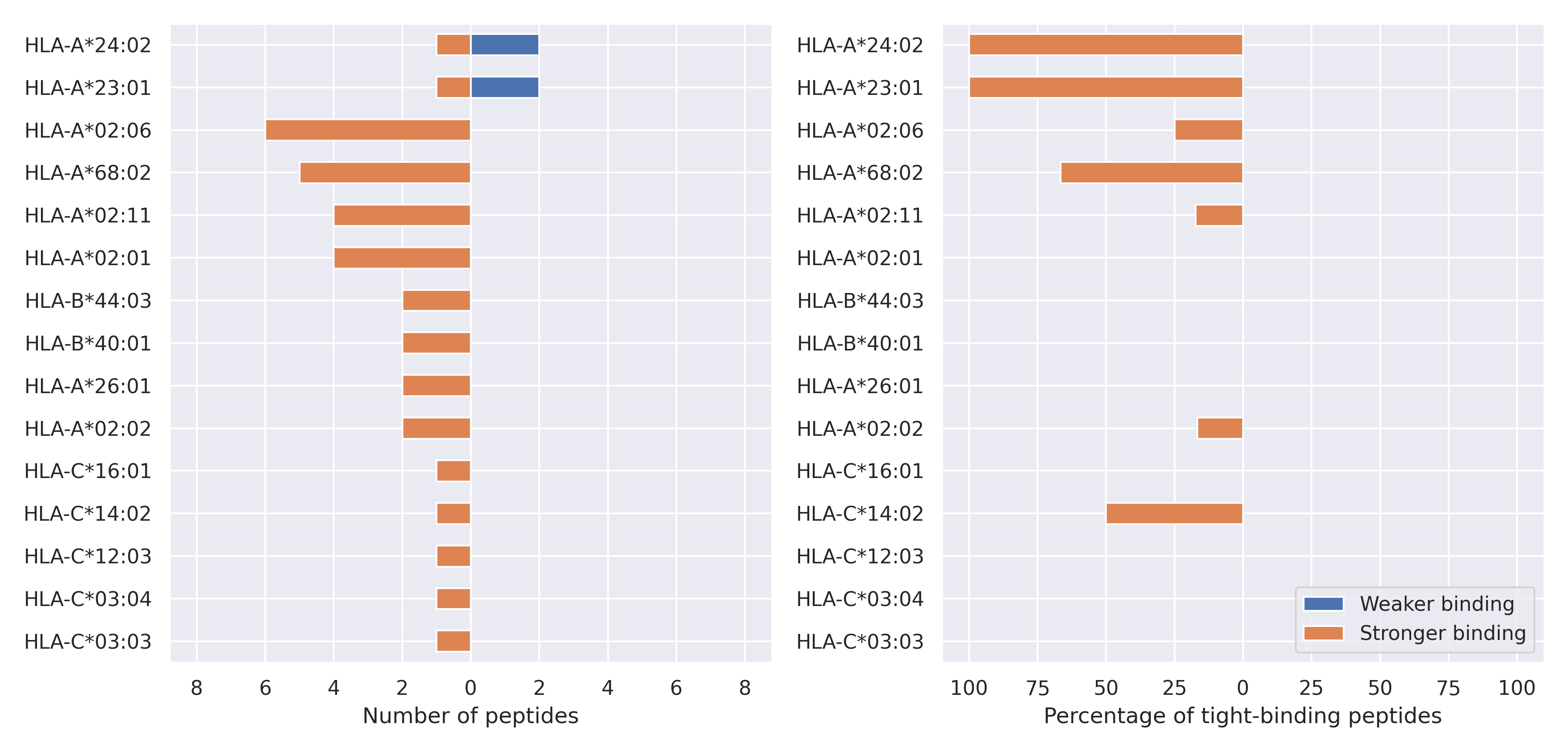
T9I
Reference MYSFVSEETGTLIVNSVLLFLAFVVFLLVTLAIL
|||||||| |||||||||||||||||||||||||
Mutated MYSFVSEEIGALIVNSVLLFLAFVVFLLVTLAIL
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-A*24:02 | MYSFVSEET | MYSFVSEEI | 5532 | 18 |
| HLA-A*23:01 | MYSFVSEET | MYSFVSEEI | 5604 | 31 |
| HLA-C*16:01 | YSFVSEET | YSFVSEEI | 7157 | 226 |
| HLA-C*14:02 | MYSFVSEET | MYSFVSEEI | 901 | 30 |
| HLA-C*12:03 | YSFVSEET | YSFVSEEI | 10273 | 457 |
| HLA-A*68:02 | FVSEETGTLI | FVSEEIGALI | 247 | 22 |
| HLA-A*68:02 | MYSFVSEET | MYSFVSEEI | 3879 | 472 |
| HLA-A*02:11 | FVSEETGTLI | FVSEEIGALI | 307 | 45 |
| HLA-A*02:01 | FVSEETGTLI | FVSEEIGALI | 1293 | 202 |
| HLA-A*02:06 | FVSEETGTLI | FVSEEIGALI | 423 | 84 |
| HLA-A*02:02 | FVSEETGTLI | FVSEEIGALI | 85 | 20 |
| HLA-A*26:01 | FVSEETGTL | FVSEEIGAL | 1352 | 323 |
| HLA-A*02:06 | FVSEETGTL | FVSEEIGAL | 120 | 30 |
| HLA-A*68:02 | YSFVSEETGTLI | YSFVSEEIGALI | 1008 | 253 |
| HLA-B*40:01 | EETGTLIVNSVL | EEIGALIVNSVL | 1578 | 420 |
| HLA-A*24:02 | MYSFVSEETGTL | MYSFVSEEIGAL | 363 | 1234 |
| HLA-A*23:01 | MYSFVSEETGTL | MYSFVSEEIGAL | 431 | 1384 |
| HLA-A*02:01 | FVSEETGTL | FVSEEIGAL | 575 | 183 |
| HLA-A*02:06 | FVSEETGTLIV | FVSEEIGALIV | 670 | 227 |
| HLA-A*02:11 | FVSEETGTL | FVSEEIGAL | 97 | 35 |
| HLA-B*44:03 | EETGTLIVNSVLLF | EEIGALIVNSVLLF | 877 | 385 |
T11A
Reference MYSFVSEETGTLIVNSVLLFLAFVVFLLVTLAILTA
|||||||||| |||||||||||||||||||||||||
Mutated MYSFVSEEIGALIVNSVLLFLAFVVFLLVTLAILTA
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-A*68:02 | FVSEETGTLI | FVSEEIGALI | 247 | 22 |
| HLA-C*03:03 | GTLIVNSVL | GALIVNSVL | 2389 | 293 |
| HLA-C*03:04 | GTLIVNSVL | GALIVNSVL | 2389 | 293 |
| HLA-A*02:11 | FVSEETGTLI | FVSEEIGALI | 307 | 45 |
| HLA-A*02:01 | FVSEETGTLI | FVSEEIGALI | 1293 | 202 |
| HLA-A*02:06 | FVSEETGTLI | FVSEEIGALI | 423 | 84 |
| HLA-A*02:02 | FVSEETGTLI | FVSEEIGALI | 85 | 20 |
| HLA-A*26:01 | FVSEETGTL | FVSEEIGAL | 1352 | 323 |
| HLA-A*02:06 | FVSEETGTL | FVSEEIGAL | 120 | 30 |
| HLA-A*68:02 | YSFVSEETGTLI | YSFVSEEIGALI | 1008 | 253 |
| HLA-B*40:01 | EETGTLIVNSVL | EEIGALIVNSVL | 1578 | 420 |
| HLA-A*24:02 | MYSFVSEETGTL | MYSFVSEEIGAL | 363 | 1234 |
| HLA-A*23:01 | MYSFVSEETGTL | MYSFVSEEIGAL | 431 | 1384 |
| HLA-A*02:01 | FVSEETGTL | FVSEEIGAL | 575 | 183 |
| HLA-A*02:06 | FVSEETGTLIV | FVSEEIGALIV | 670 | 227 |
| HLA-A*02:11 | FVSEETGTL | FVSEEIGAL | 97 | 35 |
| HLA-B*44:03 | EETGTLIVNSVLLF | EEIGALIVNSVLLF | 877 | 385 |
NS3 protein
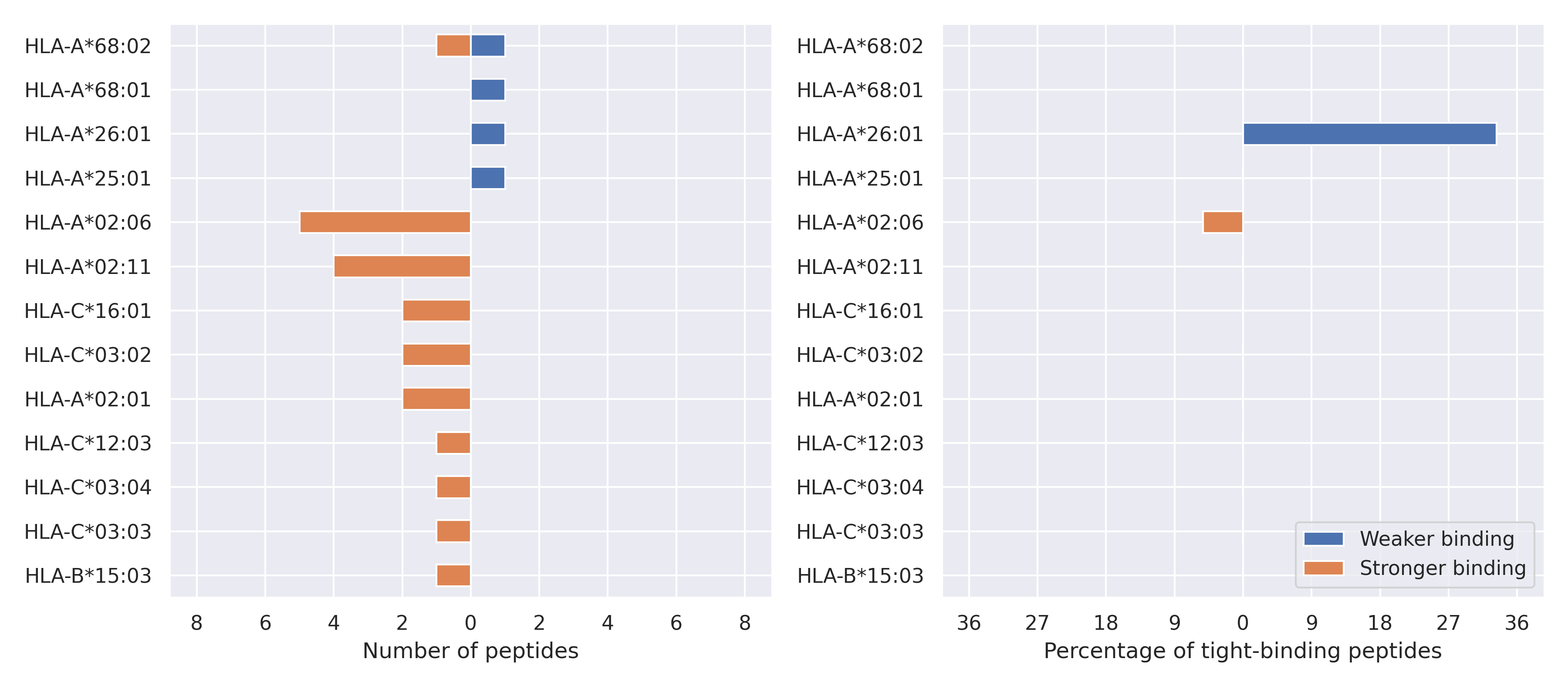
T223I
Reference KDCVVLHSYFTSDYYQLYSTQLSTDTGVEHVTFFIYNKIVDEPEEHVQIHT
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated KDCVVLHSYFTSDYYQLYSTQLSTDIGVEHVTFFIYNKIVDEPEEHVQIHT
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-C*03:03 | YSTQLSTDT | YSTQLSTDI | 12349 | 241 |
| HLA-C*03:04 | YSTQLSTDT | YSTQLSTDI | 12349 | 241 |
| HLA-C*12:03 | YSTQLSTDT | YSTQLSTDI | 11751 | 430 |
| HLA-C*16:01 | YSTQLSTDT | YSTQLSTDI | 7819 | 294 |
| HLA-C*03:02 | YSTQLSTDT | YSTQLSTDI | 6466 | 347 |
| HLA-A*02:06 | YQLYSTQLSTDT | YQLYSTQLSTDI | 4179 | 287 |
| HLA-A*02:11 | QLYSTQLSTDT | QLYSTQLSTDI | 5151 | 437 |
| HLA-A*68:01 | TGVEHVTFFIYNK | IGVEHVTFFIYNK | 213 | 1427 |
| HLA-A*68:02 | LSTDTGVEHV | LSTDIGVEHV | 1765 | 265 |
| HLA-A*68:02 | TGVEHVTFFI | IGVEHVTFFI | 212 | 1093 |
| HLA-A*02:01 | TQLSTDTGV | TQLSTDIGV | 1084 | 283 |
| HLA-A*02:01 | YSTQLSTDTGV | YSTQLSTDIGV | 1288 | 379 |
| HLA-A*02:11 | STQLSTDTGV | STQLSTDIGV | 1485 | 443 |
| HLA-A*25:01 | DTGVEHVTFF | DIGVEHVTFF | 180 | 596 |
| HLA-A*02:11 | TQLSTDTGV | TQLSTDIGV | 436 | 133 |
| HLA-A*02:06 | STQLSTDTGV | STQLSTDIGV | 264 | 83 |
| HLA-A*02:11 | YSTQLSTDTGV | YSTQLSTDIGV | 1184 | 381 |
| HLA-A*26:01 | DTGVEHVTFF | DIGVEHVTFF | 37 | 108 |
| HLA-A*02:06 | YSTQLSTDTGV | YSTQLSTDIGV | 157 | 56 |
| HLA-C*03:02 | TGVEHVTFF | IGVEHVTFF | 237 | 88 |
| HLA-A*02:06 | TQLSTDTGV | TQLSTDIGV | 70 | 26 |
| HLA-B*15:03 | TGVEHVTFF | IGVEHVTFF | 305 | 115 |
| HLA-A*02:06 | LYSTQLSTDTGV | LYSTQLSTDIGV | 855 | 330 |
| HLA-C*16:01 | TGVEHVTFF | IGVEHVTFF | 392 | 185 |
NS8 protein
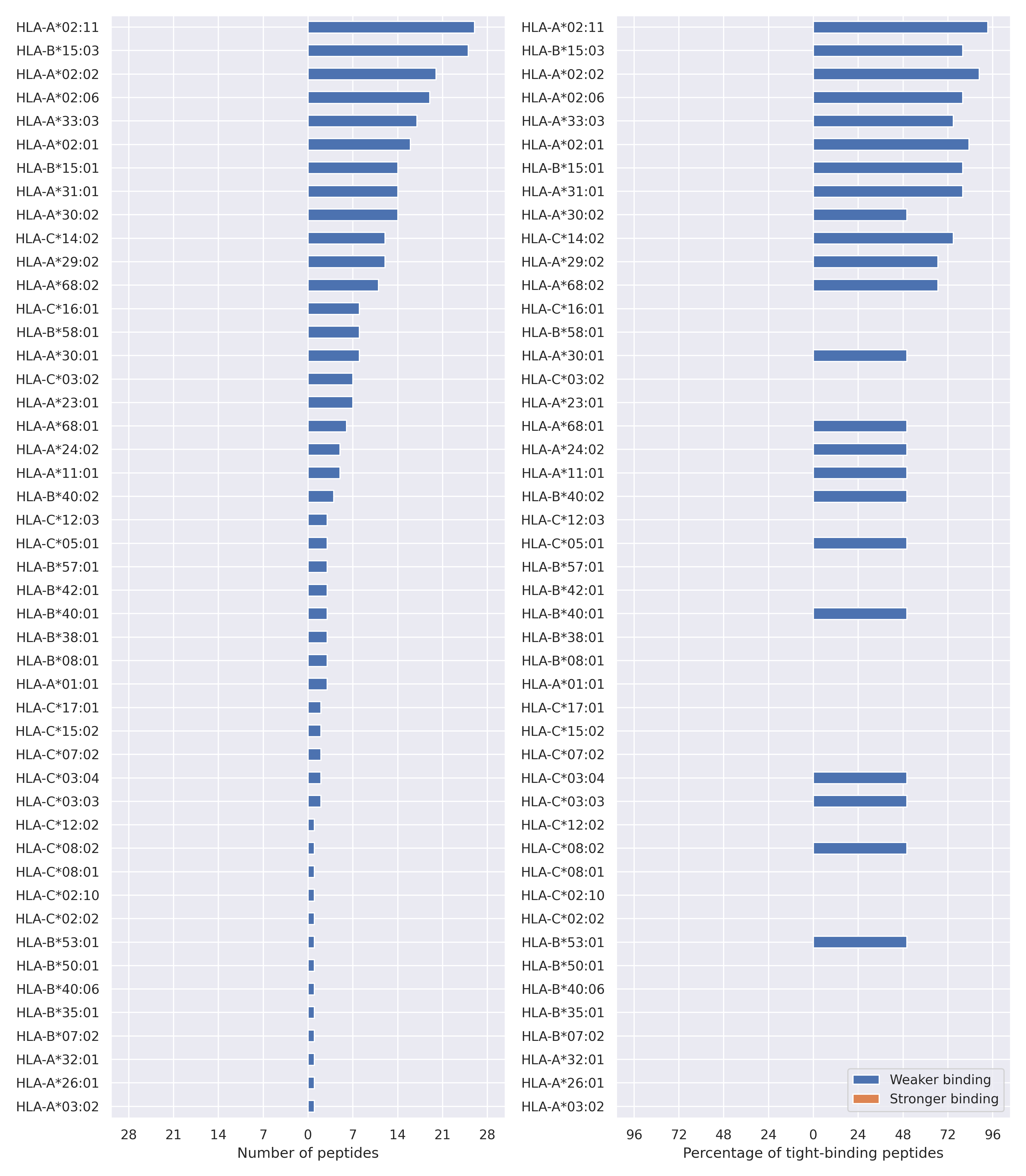
G8stop
Reference MKFLVFLGIITTVAAFHQECSLQSCTQHQPYVVDDPCPIHFYSKWYIRVGARKSAPLIELCVDEAGSKSPIQYIDIGNYTVSCLPFTINCQEPKLGSLVVRCSFYEDFLEYHDVRVVLDFI
|||||||
Mutated MKFLVFL‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-A*02:11 | YIDIGNYTV | - | 3 | - |
| HLA-A*29:02 | SFYEDFLEY | - | 3 | - |
| HLA-A*31:01 | HFYSKWYIR | - | 4 | - |
| HLA-A*02:11 | FLEYHDVRV | - | 5 | - |
| HLA-A*33:03 | HFYSKWYIR | - | 5 | - |
| HLA-A*02:06 | YIDIGNYTV | - | 6 | - |
| HLA-B*15:03 | IQYIDIGNY | - | 6 | - |
| HLA-A*02:11 | FLVFLGIITTV | FLVFL | 7 | - |
| HLA-A*68:01 | FTINCQEPK | - | 7 | - |
| HLA-A*02:01 | YIDIGNYTV | - | 8 | - |
| HLA-A*02:02 | FLEYHDVRV | - | 9 | - |
| HLA-A*02:11 | FLGIITTV | FL | 9 | - |
| HLA-A*02:06 | YVVDDPCPI | - | 10 | - |
| HLA-A*02:11 | FLEYHDVRVV | - | 10 | - |
| HLA-B*15:03 | LQSCTQHQPY | - | 10 | - |
| HLA-C*14:02 | SFYEDFLEY | - | 10 | - |
| HLA-A*02:11 | YVVDDPCPI | - | 13 | - |
| HLA-A*31:01 | IHFYSKWYIR | - | 13 | - |
| HLA-B*53:01 | CPIHFYSKW | - | 13 | - |
| HLA-C*14:02 | NYTVSCLPF | - | 13 | - |
| HLA-B*15:01 | LQSCTQHQPY | - | 14 | - |
| HLA-A*29:02 | CSFYEDFLEY | - | 15 | - |
| HLA-A*02:02 | YIDIGNYTV | - | 16 | - |
| HLA-C*05:01 | YIDIGNYTV | - | 16 | - |
| HLA-A*02:06 | YTVSCLPFTI | - | 18 | - |
| HLA-B*15:01 | LGIITTVAAF | L | 18 | - |
| HLA-B*15:01 | GIITTVAAF | - | 18 | - |
| HLA-A*68:02 | TVSCLPFTI | - | 19 | - |
| HLA-A*02:11 | FLGIITTVA | FL | 20 | - |
| HLA-A*02:01 | FLEYHDVRV | - | 22 | - |
| HLA-A*31:01 | KWYIRVGAR | - | 22 | - |
| HLA-A*02:02 | FLEYHDVRVV | - | 24 | - |
| HLA-A*68:02 | YTVSCLPFTI | - | 24 | - |
| HLA-A*02:01 | FLVFLGIITTV | FLVFL | 25 | - |
| HLA-A*02:11 | LVFLGIITTV | LVFL | 25 | - |
| HLA-A*02:02 | FLGIITTVA | FL | 27 | - |
| HLA-A*02:02 | FLGIITTV | FL | 29 | - |
| HLA-A*02:02 | YVVDDPCPI | - | 29 | - |
| HLA-A*02:11 | YTVSCLPFTI | - | 29 | - |
| HLA-C*03:03 | YVVDDPCPI | - | 29 | - |
| HLA-C*03:04 | YVVDDPCPI | - | 29 | - |
| HLA-A*11:01 | FTINCQEPK | - | 30 | - |
| HLA-C*08:02 | YIDIGNYTV | - | 30 | - |
| HLA-A*02:06 | LVFLGIITTV | LVFL | 31 | - |
| HLA-A*02:11 | SLQSCTQHQPYV | - | 31 | - |
| HLA-A*31:01 | HFYSKWYIRVGAR | - | 31 | - |
| HLA-A*02:11 | TVSCLPFTI | - | 32 | - |
| HLA-A*33:03 | HFYSKWYIRVGAR | - | 32 | - |
| HLA-B*15:01 | IQYIDIGNY | - | 32 | - |
| HLA-B*40:01 | LEYHDVRVVL | - | 32 | - |
| HLA-A*33:03 | IHFYSKWYIR | - | 35 | - |
| HLA-A*02:02 | YTVSCLPFTI | - | 36 | - |
| HLA-A*02:11 | KFLVFLGIITTV | KFLVFL | 36 | - |
| HLA-C*14:02 | EYHDVRVVL | - | 38 | - |
| HLA-A*30:02 | IQYIDIGNY | - | 39 | - |
| HLA-B*15:03 | SFYEDFLEY | - | 39 | - |
| HLA-B*15:03 | GIITTVAAF | - | 40 | - |
| HLA-A*02:01 | FLGIITTV | FL | 42 | - |
| HLA-A*02:11 | FLVFLGIITT | FLVFL | 45 | - |
| HLA-A*02:01 | FLEYHDVRVV | - | 46 | - |
| HLA-A*02:02 | FLGIITTVAA | FL | 47 | - |
| HLA-A*24:02 | NYTVSCLPF | - | 47 | - |
| HLA-A*30:01 | GSKSPIQYI | - | 47 | - |
| HLA-A*02:11 | FLGIITTVAA | FL | 48 | - |
| HLA-B*40:02 | LEYHDVRVVL | - | 49 | - |
| HLA-C*03:02 | YTVSCLPF | - | 52 | - |
| HLA-A*23:01 | NYTVSCLPF | - | 53 | - |
| HLA-A*30:02 | GSLVVRCSFY | - | 54 | - |
| HLA-A*29:02 | VRCSFYEDFLEY | - | 55 | - |
| HLA-A*30:01 | GARKSAPLI | - | 55 | - |
| HLA-B*08:01 | YIRVGARKSAPL | - | 55 | - |
| HLA-C*03:02 | YVVDDPCPI | - | 55 | - |
| HLA-A*02:11 | FLVFLGIITTVA | FLVFL | 56 | - |
| HLA-A*02:02 | FLVFLGIITTV | FLVFL | 57 | - |
| HLA-A*68:02 | YVVDDPCPI | - | 57 | - |
| HLA-A*02:01 | YVVDDPCPI | - | 59 | - |
| HLA-A*02:06 | YTVSCLPFT | - | 59 | - |
| HLA-B*15:03 | RKSAPLIEL | - | 59 | - |
| HLA-A*29:02 | RCSFYEDFLEY | - | 60 | - |
| HLA-A*29:02 | VVRCSFYEDFLEY | - | 61 | - |
| HLA-A*02:02 | TVSCLPFTI | - | 63 | - |
| HLA-A*02:06 | FLVFLGIITTV | FLVFL | 64 | - |
| HLA-C*03:03 | AAFHQECSL | - | 65 | - |
| HLA-C*03:04 | AAFHQECSL | - | 65 | - |
| HLA-A*68:02 | LVFLGIITTV | LVFL | 66 | - |
| HLA-B*15:03 | MKFLVFLGI | MKFLVFL | 73 | - |
| HLA-C*16:01 | AAFHQECSL | - | 73 | - |
| HLA-A*31:01 | CPIHFYSKWYIR | - | 74 | - |
| HLA-A*30:01 | RVGARKSAPLI | - | 75 | - |
| HLA-A*01:01 | CSFYEDFLEY | - | 76 | - |
| HLA-A*02:06 | FLEYHDVRV | - | 80 | - |
| HLA-A*02:01 | FLVFLGIITT | FLVFL | 83 | - |
| HLA-A*02:02 | FLVFLGIITT | FLVFL | 83 | - |
| HLA-A*29:02 | SLVVRCSFY | - | 84 | - |
| HLA-A*30:02 | LQSCTQHQPY | - | 84 | - |
| HLA-A*02:06 | TVSCLPFTI | - | 85 | - |
| HLA-A*02:02 | FLEYHDVRVVL | - | 86 | - |
| HLA-A*30:02 | CSFYEDFLEY | - | 89 | - |
| HLA-A*02:11 | VFLGIITTV | VFL | 90 | - |
| HLA-A*30:02 | SFYEDFLEY | - | 90 | - |
| HLA-C*14:02 | LEYHDVRVVL | - | 90 | - |
| HLA-A*02:01 | YTVSCLPFTI | - | 93 | - |
| HLA-A*02:02 | LVFLGIITTV | LVFL | 94 | - |
| HLA-B*58:01 | YTVSCLPFTI | - | 94 | - |
| HLA-A*02:01 | LVFLGIITTV | LVFL | 96 | - |
| HLA-C*12:03 | YVVDDPCPI | - | 97 | - |
| HLA-A*29:02 | YVVDDPCPIHFY | - | 98 | - |
| HLA-A*31:01 | HFYSKWYIRV | - | 98 | - |
| HLA-A*02:11 | KLGSLVVRC | - | 101 | - |
| HLA-B*15:03 | GNYTVSCLPF | - | 101 | - |
| HLA-A*30:02 | SLVVRCSFY | - | 103 | - |
| HLA-A*26:01 | YVVDDPCPIHFY | - | 106 | - |
| HLA-A*02:02 | SLQSCTQHQPYV | - | 108 | - |
| HLA-A*29:02 | SFYEDFLEYH | - | 110 | - |
| HLA-A*68:02 | FTINCQEPKL | - | 113 | - |
| HLA-B*15:01 | SLVVRCSFY | - | 115 | - |
| HLA-C*14:02 | FYEDFLEY | - | 116 | - |
| HLA-A*33:03 | DFLEYHDVR | - | 117 | - |
| HLA-A*02:01 | FLGIITTVA | FL | 118 | - |
| HLA-B*42:01 | EPKLGSLVV | - | 118 | - |
| HLA-A*31:01 | YSKWYIRVGAR | - | 120 | - |
| HLA-A*03:02 | FTINCQEPK | - | 121 | - |
| HLA-A*02:01 | FLGIITTVAA | FL | 122 | - |
| HLA-A*30:02 | KSPIQYIDIGNY | - | 122 | - |
| HLA-B*40:02 | LEYHDVRVV | - | 122 | - |
| HLA-A*33:03 | CPIHFYSKWYIR | - | 123 | - |
| HLA-B*15:03 | LGIITTVAAF | L | 123 | - |
| HLA-C*14:02 | FLEYHDVRVVL | - | 126 | - |
| HLA-B*15:03 | CSFYEDFLEY | - | 128 | - |
| HLA-B*35:01 | SFYEDFLEY | - | 128 | - |
| HLA-A*68:01 | PFTINCQEPK | - | 130 | - |
| HLA-B*15:03 | SLVVRCSFY | - | 130 | - |
| HLA-B*15:01 | FLGIITTVAAF | FL | 131 | - |
| HLA-A*33:03 | FYSKWYIRVGAR | - | 133 | - |
| HLA-B*15:03 | GSLVVRCSF | - | 133 | - |
| HLA-A*02:06 | FLGIITTV | FL | 134 | - |
| HLA-A*02:06 | FTINCQEPKL | - | 134 | - |
| HLA-A*02:11 | QYIDIGNYTV | - | 135 | - |
| HLA-A*02:11 | FLEYHDVRVVL | - | 137 | - |
| HLA-A*02:01 | KFLVFLGIITTV | KFLVFL | 138 | - |
| HLA-B*57:01 | LGSLVVRCSF | - | 138 | - |
| HLA-A*02:02 | FTINCQEPKL | - | 140 | - |
| HLA-A*31:01 | KWYIRVGARK | - | 145 | - |
| HLA-A*68:01 | HFYSKWYIR | - | 146 | - |
| HLA-A*30:01 | KWYIRVGARK | - | 149 | - |
| HLA-C*03:02 | SFYEDFLEY | - | 152 | - |
| HLA-C*12:02 | YVVDDPCPI | - | 156 | - |
| HLA-A*31:01 | PIHFYSKWYIR | - | 158 | - |
| HLA-B*15:03 | YTVSCLPF | - | 158 | - |
| HLA-C*07:02 | SFYEDFLEY | - | 160 | - |
| HLA-A*33:03 | YSKWYIRVGAR | - | 162 | - |
| HLA-C*17:01 | YVVDDPCPI | - | 163 | - |
| HLA-A*02:01 | SLQSCTQHQPYV | - | 164 | - |
| HLA-B*38:01 | IHFYSKWYI | - | 168 | - |
| HLA-A*23:01 | FYSKWYIRV | - | 171 | - |
| HLA-A*29:02 | LVVRCSFYEDFLEY | - | 171 | - |
| HLA-A*02:11 | SLQSCTQHQPYVV | - | 172 | - |
| HLA-A*23:01 | QYIDIGNYTV | - | 174 | - |
| HLA-A*33:03 | FYSKWYIR | - | 174 | - |
| HLA-A*33:03 | HFYSKWYIRV | - | 176 | - |
| HLA-B*58:01 | KSAPLIELCV | - | 177 | - |
| HLA-A*23:01 | VFLGIITTVAAF | VFL | 180 | - |
| HLA-C*17:01 | YIDIGNYTV | - | 181 | - |
| HLA-B*58:01 | LGSLVVRCSF | - | 182 | - |
| HLA-A*31:01 | SKWYIRVGAR | - | 183 | - |
| HLA-A*02:01 | FLVFLGIITTVA | FLVFL | 184 | - |
| HLA-B*38:01 | YHDVRVVLDF | - | 184 | - |
| HLA-C*14:02 | VFLGIITTV | VFL | 189 | - |
| HLA-A*02:06 | FLEYHDVRVV | - | 192 | - |
| HLA-A*30:01 | AGSKSPIQYI | - | 192 | - |
| HLA-C*03:02 | AAFHQECSL | - | 192 | - |
| HLA-C*08:01 | YIDIGNYTV | - | 192 | - |
| HLA-C*12:03 | AAFHQECSL | - | 192 | - |
| HLA-A*11:01 | SCLPFTINCQEPK | - | 193 | - |
| HLA-A*29:02 | FYEDFLEY | - | 195 | - |
| HLA-A*31:01 | FYSKWYIR | - | 195 | - |
| HLA-A*30:02 | KLGSLVVRCSFY | - | 199 | - |
| HLA-B*15:03 | SPIQYIDIGNY | - | 201 | - |
| HLA-B*58:01 | GSLVVRCSF | - | 201 | - |
| HLA-A*02:11 | MKFLVFLGIITTV | MKFLVFL | 205 | - |
| HLA-C*14:02 | FYSKWYIRV | - | 207 | - |
| HLA-B*58:01 | CSFYEDFLEY | - | 208 | - |
| HLA-C*03:02 | CSFYEDFLEY | - | 211 | - |
| HLA-A*02:02 | KFLVFLGIITTV | KFLVFL | 215 | - |
| HLA-B*58:01 | KSAPLIELC | - | 217 | - |
| HLA-B*42:01 | QEPKLGSLVV | - | 218 | - |
| HLA-B*15:01 | LVFLGIITTVAAF | LVFL | 219 | - |
| HLA-A*24:02 | NYTVSCLPFTI | - | 221 | - |
| HLA-B*50:01 | LEYHDVRVV | - | 221 | - |
| HLA-B*40:01 | YEDFLEYHDVRVVL | - | 224 | - |
| HLA-A*01:01 | VVDDPCPIHFY | - | 225 | - |
| HLA-B*07:02 | RVGARKSAPL | - | 225 | - |
| HLA-A*30:01 | YSKWYIRVGA | - | 229 | - |
| HLA-C*16:01 | YVVDDPCPI | - | 229 | - |
| HLA-A*23:01 | NYTVSCLPFTI | - | 231 | - |
| HLA-A*02:06 | FLVFLGIITT | FLVFL | 234 | - |
| HLA-A*30:02 | RCSFYEDFLEY | - | 236 | - |
| HLA-A*68:02 | DVRVVLDFI | - | 236 | - |
| HLA-A*33:03 | DPCPIHFYSKWYIR | - | 238 | - |
| HLA-C*16:01 | YTVSCLPF | - | 240 | - |
| HLA-A*02:01 | TVSCLPFTI | - | 244 | - |
| HLA-A*31:01 | FYSKWYIRVGAR | - | 244 | - |
| HLA-C*05:01 | FLEYHDVRV | - | 244 | - |
| HLA-A*68:02 | YTVSCLPFT | - | 245 | - |
| HLA-C*05:01 | VVDDPCPIHF | - | 247 | - |
| HLA-A*02:11 | FLVFLGIITTVAA | FLVFL | 248 | - |
| HLA-C*16:01 | VAAFHQECSL | - | 251 | - |
| HLA-B*15:03 | KSPIQYIDIGNY | - | 252 | - |
| HLA-A*68:02 | FTINCQEPKLGSLV | - | 254 | - |
| HLA-B*15:01 | CSFYEDFLEY | - | 256 | - |
| HLA-A*24:02 | FYSKWYIRV | - | 261 | - |
| HLA-A*68:02 | YSKWYIRVGA | - | 262 | - |
| HLA-A*02:02 | FLVFLGIITTVA | FLVFL | 263 | - |
| HLA-B*57:01 | KLGSLVVRCSF | - | 263 | - |
| HLA-A*01:01 | CVDEAGSKSPIQY | - | 264 | - |
| HLA-A*33:03 | EDFLEYHDVR | - | 264 | - |
| HLA-A*23:01 | IGNYTVSCLPF | - | 272 | - |
| HLA-B*15:03 | SLQSCTQHQPY | - | 275 | - |
| HLA-C*02:02 | YVVDDPCPI | - | 275 | - |
| HLA-C*02:10 | YVVDDPCPI | - | 275 | - |
| HLA-B*57:01 | GSLVVRCSF | - | 276 | - |
| HLA-A*02:06 | FLGIITTVAA | FL | 282 | - |
| HLA-A*68:02 | TTVAAFHQECSL | - | 288 | - |
| HLA-A*32:01 | TVSCLPFTI | - | 294 | - |
| HLA-A*02:02 | SLVVRCSFYEDFL | - | 298 | - |
| HLA-A*02:11 | IQYIDIGNYTV | - | 299 | - |
| HLA-A*30:02 | PIQYIDIGNY | - | 304 | - |
| HLA-A*24:02 | QYIDIGNYTV | - | 310 | - |
| HLA-A*02:11 | YIDIGNYTVS | - | 313 | - |
| HLA-A*02:06 | IQYIDIGNYTV | - | 320 | - |
| HLA-B*15:01 | VVRCSFYEDF | - | 326 | - |
| HLA-B*40:01 | LEYHDVRVV | - | 328 | - |
| HLA-B*15:03 | RCSFYEDFLEY | - | 332 | - |
| HLA-B*15:01 | SLQSCTQHQPY | - | 333 | - |
| HLA-C*14:02 | YEDFLEYHDVRVVL | - | 333 | - |
| HLA-A*68:01 | YVVDDPCPIHFYSK | - | 335 | - |
| HLA-A*02:02 | VFLGIITTVA | VFL | 336 | - |
| HLA-A*31:01 | IHFYSKWYIRVGAR | - | 336 | - |
| HLA-C*03:02 | GIITTVAAF | - | 338 | - |
| HLA-B*08:01 | VGARKSAPL | - | 346 | - |
| HLA-C*14:02 | YHDVRVVL | - | 346 | - |
| HLA-A*33:03 | KWYIRVGAR | - | 347 | - |
| HLA-B*15:03 | QSCTQHQPY | - | 350 | - |
| HLA-B*15:01 | SPIQYIDIGNY | - | 352 | - |
| HLA-B*40:02 | YEDFLEYHDV | - | 352 | - |
| HLA-A*33:03 | WYIRVGARK | - | 359 | - |
| HLA-A*33:03 | SKWYIRVGAR | - | 361 | - |
| HLA-C*03:02 | YVVDDPCPIHF | - | 364 | - |
| HLA-A*02:06 | LVFLGIITT | LVFL | 366 | - |
| HLA-A*02:02 | SLQSCTQHQPYVV | - | 376 | - |
| HLA-A*02:06 | KFLVFLGIITTV | KFLVFL | 382 | - |
| HLA-A*30:01 | KWYIRVGAR | - | 382 | - |
| HLA-C*16:01 | IGNYTVSCL | - | 383 | - |
| HLA-B*15:03 | SKSPIQYIDIGNY | - | 384 | - |
| HLA-A*30:02 | QSCTQHQPY | - | 386 | - |
| HLA-B*15:03 | PIQYIDIGNY | - | 390 | - |
| HLA-A*02:06 | FLGIITTVA | FL | 391 | - |
| HLA-A*30:02 | SLQSCTQHQPY | - | 393 | - |
| HLA-A*11:01 | CSFYEDFLEY | - | 395 | - |
| HLA-A*68:01 | FTINCQEPKL | - | 395 | - |
| HLA-C*16:01 | QSCTQHQPY | - | 397 | - |
| HLA-A*33:03 | PIHFYSKWYIR | - | 398 | - |
| HLA-B*08:01 | SLVVRCSF | - | 399 | - |
| HLA-B*15:03 | AAFHQECSL | - | 400 | - |
| HLA-C*16:01 | GSLVVRCSF | - | 400 | - |
| HLA-A*11:01 | VVDDPCPIHFYSK | - | 401 | - |
| HLA-A*29:02 | GSLVVRCSFY | - | 402 | - |
| HLA-A*68:01 | YSKWYIRVGARK | - | 402 | - |
| HLA-C*15:02 | KSAPLIELCV | - | 403 | - |
| HLA-C*07:02 | EYHDVRVVL | - | 404 | - |
| HLA-A*29:02 | IQYIDIGNY | - | 410 | - |
| HLA-A*02:11 | YIDIGNYTVSCL | - | 412 | - |
| HLA-B*40:06 | LEYHDVRVV | - | 412 | - |
| HLA-A*68:02 | YIDIGNYTV | - | 413 | - |
| HLA-B*15:03 | YHDVRVVLDF | - | 413 | - |
| HLA-A*33:03 | FTINCQEPK | - | 416 | - |
| HLA-B*38:01 | YHDVRVVL | - | 417 | - |
| HLA-A*24:02 | IGNYTVSCLPF | - | 426 | - |
| HLA-A*30:01 | HFYSKWYIR | - | 430 | - |
| HLA-C*15:02 | YTVSCLPFTI | - | 430 | - |
| HLA-B*15:03 | CQEPKLGSL | - | 432 | - |
| HLA-B*15:01 | VFLGIITTVAAF | VFL | 439 | - |
| HLA-C*12:03 | SFYEDFLEY | - | 443 | - |
| HLA-B*42:01 | YIRVGARKSAPL | - | 448 | - |
| HLA-B*15:03 | VRCSFYEDFLEY | - | 449 | - |
| HLA-B*58:01 | KLGSLVVRCSF | - | 450 | - |
| HLA-C*14:02 | CSFYEDFLEY | - | 454 | - |
| HLA-B*15:01 | YTVSCLPF | - | 456 | - |
| HLA-A*30:02 | SKSPIQYIDIGNY | - | 458 | - |
| HLA-A*02:11 | DFLEYHDVRVV | - | 459 | - |
| HLA-A*11:01 | LPFTINCQEPK | - | 466 | - |
| HLA-B*40:02 | YEDFLEYHDVRVVL | - | 468 | - |
| HLA-B*15:03 | IQYIDIGNYT | - | 483 | - |
| HLA-B*15:03 | FLGIITTVAAF | FL | 484 | - |
| HLA-B*58:01 | TVSCLPFTI | - | 484 | - |
| HLA-B*15:01 | FLVFLGIITTVAAF | FLVFL | 485 | - |
| HLA-C*16:01 | SFYEDFLEY | - | 486 | - |
| HLA-A*30:02 | SPIQYIDIGNY | - | 491 | - |
| HLA-A*02:06 | FTINCQEPKLGSL | - | 492 | - |
| HLA-A*23:01 | GNYTVSCLPF | - | 495 | - |
| HLA-A*33:03 | IHFYSKWYIRVGAR | - | 497 | - |
| HLA-C*14:02 | FYEDFLEYH | - | 497 | - |
| HLA-A*02:01 | FLEYHDVRVVL | - | 499 | - |
| HLA-A*02:06 | LQSCTQHQPYV | - | 500 | - |
| HLA-A*31:01 | WYIRVGARK | - | 500 | - |
NS9b protein
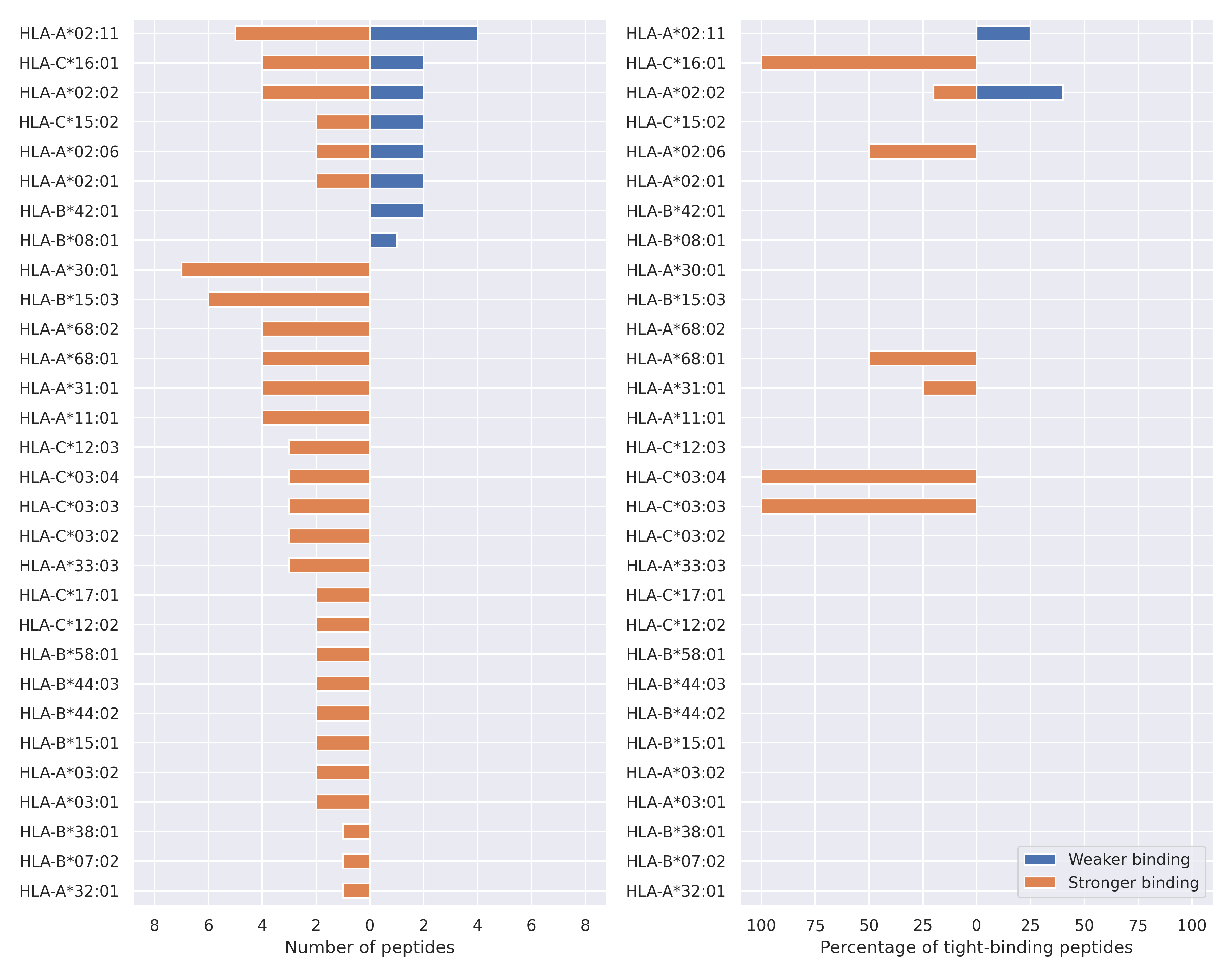
I5T
Reference MDPKISEMHPALRLVDPQIQLAVTRMENAV
|||| |||||||||||||||||||||||||
Mutated MDPKTSEMHSALRLVDPQIQLAVTRMVNAV
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-A*68:02 | MDPKISEMHPAL | MDPKTSEMHSAL | 6619 | 217 |
| HLA-A*02:01 | KISEMHPAL | KTSEMHSAL | 139 | 3161 |
| HLA-A*02:11 | KISEMHPAL | KTSEMHSAL | 14 | 277 |
| HLA-B*58:01 | KISEMHPAL | KTSEMHSAL | 5915 | 431 |
| HLA-C*15:02 | KISEMHPAL | KTSEMHSAL | 746 | 60 |
| HLA-C*16:01 | KISEMHPAL | KTSEMHSAL | 189 | 18 |
| HLA-C*12:03 | KISEMHPAL | KTSEMHSAL | 1242 | 122 |
| HLA-A*68:01 | ISEMHPALR | TSEMHSALR | 372 | 42 |
| HLA-A*02:06 | KISEMHPAL | KTSEMHSAL | 62 | 376 |
| HLA-C*12:02 | KISEMHPAL | KTSEMHSAL | 2061 | 349 |
| HLA-C*03:03 | KISEMHPAL | KTSEMHSAL | 177 | 34 |
| HLA-C*03:04 | KISEMHPAL | KTSEMHSAL | 177 | 34 |
| HLA-A*68:01 | KISEMHPALR | KTSEMHSALR | 312 | 67 |
| HLA-A*02:02 | KISEMHPAL | KTSEMHSAL | 42 | 187 |
| HLA-C*03:02 | KISEMHPAL | KTSEMHSAL | 371 | 92 |
| HLA-B*44:02 | ISEMHPALRL | TSEMHSALRL | 493 | 128 |
| HLA-A*11:01 | KISEMHPALR | KTSEMHSALR | 471 | 125 |
| HLA-B*44:03 | ISEMHPALRL | TSEMHSALRL | 577 | 160 |
| HLA-A*30:01 | KISEMHPAL | KTSEMHSAL | 455 | 143 |
| HLA-C*16:01 | ISEMHPAL | TSEMHSAL | 248 | 665 |
| HLA-A*33:03 | ISEMHPALR | TSEMHSALR | 395 | 148 |
| HLA-A*02:11 | KISEMHPALRLV | KTSEMHSALRLV | 249 | 617 |
| HLA-A*30:01 | KISEMHPALRLV | KTSEMHSALRLV | 722 | 292 |
| HLA-C*15:02 | ISEMHPALRL | TSEMHSALRL | 480 | 1127 |
| HLA-A*31:01 | PKISEMHPALR | PKTSEMHSALR | 687 | 297 |
| HLA-C*17:01 | KISEMHPAL | KTSEMHSAL | 241 | 105 |
P10S
Reference MDPKISEMHPALRLVDPQIQLAVTRMENAVGRDQN
||||||||| |||||||||||||||||||||||||
Mutated MDPKTSEMHSALRLVDPQIQLAVTRMVNAVGRDQN
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-B*42:01 | HPALRLVDPQIQL | HSALRLVDPQIQL | 148 | 15057 |
| HLA-B*42:01 | HPALRLVDPQI | HSALRLVDPQI | 297 | 15937 |
| HLA-A*68:02 | MDPKISEMHPAL | MDPKTSEMHSAL | 6619 | 217 |
| HLA-A*02:01 | KISEMHPAL | KTSEMHSAL | 139 | 3161 |
| HLA-A*02:11 | KISEMHPAL | KTSEMHSAL | 14 | 277 |
| HLA-B*58:01 | KISEMHPAL | KTSEMHSAL | 5915 | 431 |
| HLA-C*15:02 | KISEMHPAL | KTSEMHSAL | 746 | 60 |
| HLA-A*02:11 | PALRLVDPQIQL | SALRLVDPQIQL | 3683 | 325 |
| HLA-C*16:01 | KISEMHPAL | KTSEMHSAL | 189 | 18 |
| HLA-C*12:03 | KISEMHPAL | KTSEMHSAL | 1242 | 122 |
| HLA-A*68:01 | ISEMHPALR | TSEMHSALR | 372 | 42 |
| HLA-B*38:01 | MHPALRLVDPQI | MHSALRLVDPQI | 3064 | 395 |
| HLA-A*02:06 | KISEMHPAL | KTSEMHSAL | 62 | 376 |
| HLA-C*12:02 | KISEMHPAL | KTSEMHSAL | 2061 | 349 |
| HLA-A*02:02 | PALRLVDPQIQL | SALRLVDPQIQL | 2220 | 388 |
| HLA-C*03:03 | KISEMHPAL | KTSEMHSAL | 177 | 34 |
| HLA-C*03:04 | KISEMHPAL | KTSEMHSAL | 177 | 34 |
| HLA-A*68:01 | KISEMHPALR | KTSEMHSALR | 312 | 67 |
| HLA-A*02:02 | KISEMHPAL | KTSEMHSAL | 42 | 187 |
| HLA-C*03:02 | KISEMHPAL | KTSEMHSAL | 371 | 92 |
| HLA-B*44:02 | ISEMHPALRL | TSEMHSALRL | 493 | 128 |
| HLA-A*11:01 | KISEMHPALR | KTSEMHSALR | 471 | 125 |
| HLA-B*44:03 | ISEMHPALRL | TSEMHSALRL | 577 | 160 |
| HLA-A*30:01 | KISEMHPAL | KTSEMHSAL | 455 | 143 |
| HLA-C*16:01 | ISEMHPAL | TSEMHSAL | 248 | 665 |
| HLA-A*33:03 | ISEMHPALR | TSEMHSALR | 395 | 148 |
| HLA-A*02:11 | KISEMHPALRLV | KTSEMHSALRLV | 249 | 617 |
| HLA-A*30:01 | KISEMHPALRLV | KTSEMHSALRLV | 722 | 292 |
| HLA-C*15:02 | ISEMHPALRL | TSEMHSALRL | 480 | 1127 |
| HLA-A*31:01 | PKISEMHPALR | PKTSEMHSALR | 687 | 297 |
| HLA-C*17:01 | KISEMHPAL | KTSEMHSAL | 241 | 105 |
| HLA-B*15:03 | SEMHPALRL | SEMHSALRL | 493 | 245 |
E27V
Reference DPKISEMHPALRLVDPQIQLAVTRMENAVGRDQNNVGPKVYPIILRLGSPL
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated DPKTSEMHSALRLVDPQIQLAVTRMVNAVGRDQNNVGPKVYPIILRLGSPL
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-A*02:11 | IQLAVTRME | IQLAVTRMV | 23336 | 70 |
| HLA-A*02:06 | IQLAVTRME | IQLAVTRMV | 9694 | 47 |
| HLA-A*02:01 | IQLAVTRME | IQLAVTRMV | 25623 | 463 |
| HLA-A*68:02 | MENAVGRDQNNV | MVNAVGRDQNNV | 13036 | 259 |
| HLA-B*15:03 | IQLAVTRME | IQLAVTRMV | 5099 | 183 |
| HLA-A*02:06 | AVTRMENAV | AVTRMVNAV | 1575 | 114 |
| HLA-A*02:11 | AVTRMENAV | AVTRMVNAV | 2404 | 180 |
| HLA-A*02:02 | RMENAVGRDQNNV | RMVNAVGRDQNNV | 3078 | 283 |
| HLA-A*02:11 | RMENAVGRDQNNV | RMVNAVGRDQNNV | 1928 | 188 |
| HLA-A*68:02 | AVTRMENAV | AVTRMVNAV | 2310 | 404 |
| HLA-A*02:02 | AVTRMENAV | AVTRMVNAV | 1454 | 300 |
| HLA-A*30:01 | VTRMENAVG | VTRMVNAVG | 395 | 82 |
| HLA-A*30:01 | AVTRMENAVG | AVTRMVNAVG | 1413 | 315 |
| HLA-A*31:01 | RMENAVGR | RMVNAVGR | 992 | 275 |
| HLA-A*31:01 | VTRMENAVGR | VTRMVNAVGR | 113 | 34 |
| HLA-A*33:03 | VTRMENAVGR | VTRMVNAVGR | 218 | 89 |
N55S
Reference VGRDQNNVGPKVYPIILRLGSPLSLNMARKTLNSLEDKAFQLTPIAVQMTK
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated VGRDQNNVGPKVYPIILRLGSPLSLSMARKTLNSLEDKAFQLTPIAVQMTK
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-C*16:01 | LNMARKTL | LSMARKTL | 6455 | 93 |
| HLA-A*03:01 | NMARKTLNSLEDK | SMARKTLNSLEDK | 2773 | 186 |
| HLA-A*03:02 | NMARKTLNSLEDK | SMARKTLNSLEDK | 3703 | 425 |
| HLA-A*30:01 | NMARKTLNSL | SMARKTLNSL | 2051 | 341 |
| HLA-C*03:03 | LGSPLSLNM | LGSPLSLSM | 699 | 121 |
| HLA-C*03:04 | LGSPLSLNM | LGSPLSLSM | 699 | 121 |
| HLA-A*02:11 | NMARKTLNSL | SMARKTLNSL | 388 | 69 |
| HLA-B*15:01 | NMARKTLNSL | SMARKTLNSL | 1233 | 246 |
| HLA-C*03:02 | LGSPLSLNM | LGSPLSLSM | 333 | 71 |
| HLA-B*15:01 | RLGSPLSLNM | RLGSPLSLSM | 257 | 56 |
| HLA-B*15:03 | LGSPLSLNM | LGSPLSLSM | 515 | 122 |
| HLA-B*15:03 | RLGSPLSLNM | RLGSPLSLSM | 483 | 116 |
| HLA-B*15:03 | NMARKTLNSL | SMARKTLNSL | 1813 | 443 |
| HLA-C*16:01 | LGSPLSLNM | LGSPLSLSM | 219 | 55 |
| HLA-A*02:01 | NMARKTLNSL | SMARKTLNSL | 1400 | 368 |
| HLA-A*03:01 | SPLSLNMARK | SPLSLSMARK | 305 | 81 |
| HLA-B*07:02 | NMARKTLNSL | SMARKTLNSL | 720 | 196 |
| HLA-A*02:02 | NMARKTLNSL | SMARKTLNSL | 76 | 26 |
| HLA-A*03:02 | SPLSLNMARK | SPLSLSMARK | 326 | 112 |
| HLA-C*12:03 | LGSPLSLNM | LGSPLSLSM | 1131 | 394 |
| HLA-B*15:03 | LRLGSPLSLNM | LRLGSPLSLSM | 1269 | 463 |
| HLA-A*11:01 | SPLSLNMARK | SPLSLSMARK | 590 | 232 |
| HLA-A*11:01 | GSPLSLNMARK | GSPLSLSMARK | 943 | 395 |
| HLA-A*32:01 | RLGSPLSLNM | RLGSPLSLSM | 998 | 488 |
| HLA-B*08:01 | SLNMARKTL | SLSMARKTL | 454 | 925 |
NS9c protein
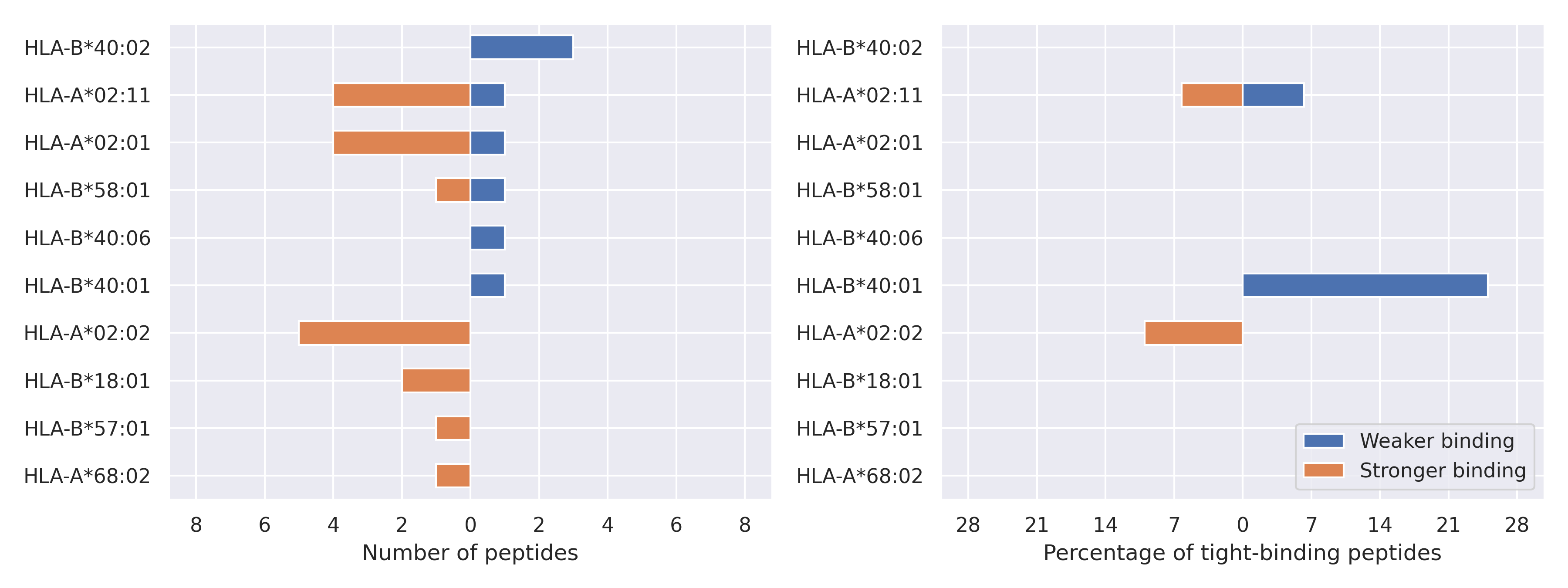
G50N
Reference AAVKPLLVPHHVVATVQEIQLQAAVGELLLLEWLAMAVMLLLLCCCLTD
||||||||||||||||||||||||| |||||||||||||||||||||||
Mutated AAVKPLLVPHHVVATVQEIQLQAAVNELLLLEWLAMAVMLLLLCCCLTD
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-B*18:01 | GELLLLEWLAM | NELLLLEWLAM | 4979 | 247 |
| HLA-B*18:01 | GELLLLEW | NELLLLEW | 3915 | 401 |
| HLA-B*40:01 | GELLLLEWL | NELLLLEWL | 28 | 140 |
| HLA-A*02:11 | QLQAAVGEL | QLQAAVNEL | 120 | 27 |
| HLA-A*02:11 | IQLQAAVGEL | IQLQAAVNEL | 338 | 81 |
| HLA-B*40:02 | GELLLLEWL | NELLLLEWL | 52 | 204 |
| HLA-B*40:02 | GELLLLEWLAM | NELLLLEWLAM | 201 | 772 |
| HLA-A*02:01 | QLQAAVGEL | QLQAAVNEL | 1290 | 358 |
| HLA-A*02:02 | QLQAAVGEL | QLQAAVNEL | 92 | 28 |
| HLA-B*58:01 | VGELLLLEW | VNELLLLEW | 330 | 1071 |
| HLA-A*02:02 | IQLQAAVGEL | IQLQAAVNEL | 170 | 58 |
| HLA-A*02:01 | LQAAVGELL | LQAAVNELL | 938 | 352 |
| HLA-A*02:02 | LQAAVGELL | LQAAVNELL | 44 | 17 |
| HLA-B*40:06 | GELLLLEWLA | NELLLLEWLA | 268 | 677 |
| HLA-B*40:02 | GELLLLEWLA | NELLLLEWLA | 475 | 1195 |
| HLA-A*02:11 | LQAAVGELL | LQAAVNELL | 386 | 154 |
| HLA-A*02:01 | IQLQAAVGEL | IQLQAAVNEL | 1138 | 475 |
| HLA-A*02:01 | QLQAAVGELL | QLQAAVNELL | 961 | 403 |
| HLA-A*02:01 | GELLLLEWLAMA | NELLLLEWLAMA | 109 | 258 |
| HLA-A*02:02 | QLQAAVGELL | QLQAAVNELL | 35 | 15 |
| HLA-A*02:02 | IQLQAAVGELL | IQLQAAVNELL | 724 | 313 |
| HLA-A*02:11 | QLQAAVGELL | QLQAAVNELL | 303 | 131 |
| HLA-B*58:01 | AVGELLLLEW | AVNELLLLEW | 196 | 87 |
| HLA-A*02:11 | GELLLLEWLAMA | NELLLLEWLAMA | 29 | 61 |
| HLA-B*57:01 | AVGELLLLEW | AVNELLLLEW | 432 | 206 |
| HLA-A*68:02 | EIQLQAAVGELL | EIQLQAAVNELL | 697 | 346 |
NSP1 protein
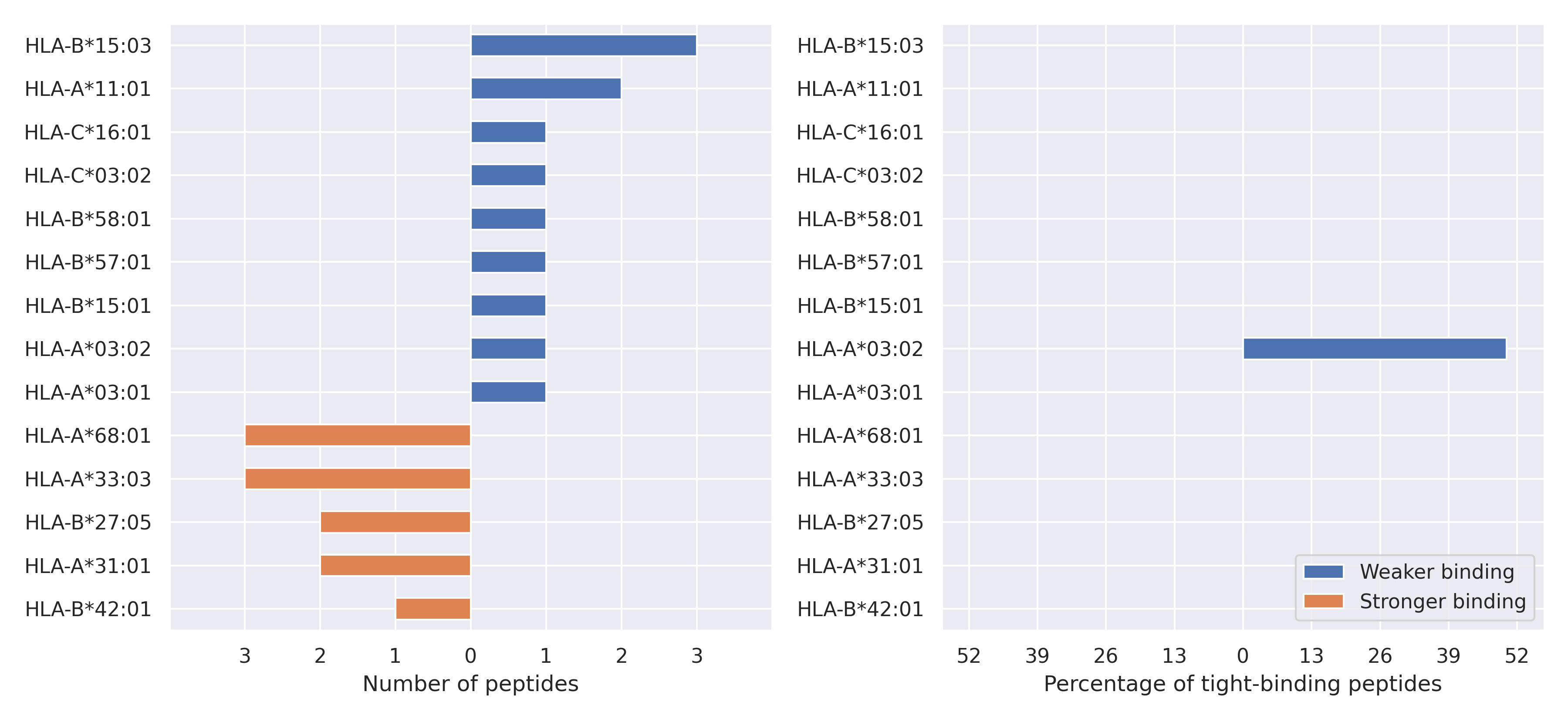
K47R
Reference QVRDVLVRGFGDSVEEVLSEARQHLKDGTCGLVEVEKGVLPQLEQPYVFIK
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated QVRDVLVRGFGDSVEEVLSEARQHLRDGTCGLVEVEKGVLPQLEQPYVFIK
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-A*03:01 | VLSEARQHLK | VLSEARQHLR | 53 | 1609 |
| HLA-A*33:03 | VLSEARQHLK | VLSEARQHLR | 5493 | 188 |
| HLA-A*33:03 | EVLSEARQHLK | EVLSEARQHLR | 4779 | 180 |
| HLA-A*33:03 | LSEARQHLK | LSEARQHLR | 5690 | 242 |
| HLA-A*11:01 | VLSEARQHLK | VLSEARQHLR | 120 | 2107 |
| HLA-A*31:01 | VLSEARQHLK | VLSEARQHLR | 1630 | 95 |
| HLA-A*31:01 | LSEARQHLK | LSEARQHLR | 1851 | 132 |
| HLA-A*11:01 | LSEARQHLK | LSEARQHLR | 223 | 2597 |
| HLA-A*03:02 | VLSEARQHLK | VLSEARQHLR | 46 | 397 |
| HLA-B*42:01 | HLKDGTCGL | HLRDGTCGL | 1455 | 390 |
| HLA-A*68:01 | LSEARQHLK | LSEARQHLR | 1065 | 302 |
| HLA-A*68:01 | VLSEARQHLK | VLSEARQHLR | 1008 | 287 |
| HLA-A*68:01 | EVLSEARQHLK | EVLSEARQHLR | 361 | 119 |
S135R
Reference HVGEIPVAYRKVLLRKNGNKGAGGHSYGADLKSFDLGDELGTDPYEDFQEN
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated HVGEIPVAYRKVLLRKNGNKGAGGHRYGADLKSFDLGDELGTDPYEDFQEN
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-B*58:01 | HSYGADLKSF | HRYGADLKSF | 59 | 18326 |
| HLA-B*27:05 | HSYGADLKSF | HRYGADLKSF | 15851 | 60 |
| HLA-B*57:01 | HSYGADLKSF | HRYGADLKSF | 158 | 23790 |
| HLA-B*27:05 | HSYGADLKSFDL | HRYGADLKSFDL | 23268 | 171 |
| HLA-B*15:01 | HSYGADLKSF | HRYGADLKSF | 94 | 6548 |
| HLA-C*03:02 | HSYGADLKSF | HRYGADLKSF | 94 | 6500 |
| HLA-C*16:01 | HSYGADLKSF | HRYGADLKSF | 329 | 14251 |
| HLA-B*15:03 | RKNGNKGAGGHSY | RKNGNKGAGGHRY | 183 | 1186 |
| HLA-B*15:03 | NKGAGGHSY | NKGAGGHRY | 129 | 774 |
| HLA-B*15:03 | GNKGAGGHSY | GNKGAGGHRY | 478 | 2611 |
NSP3 protein
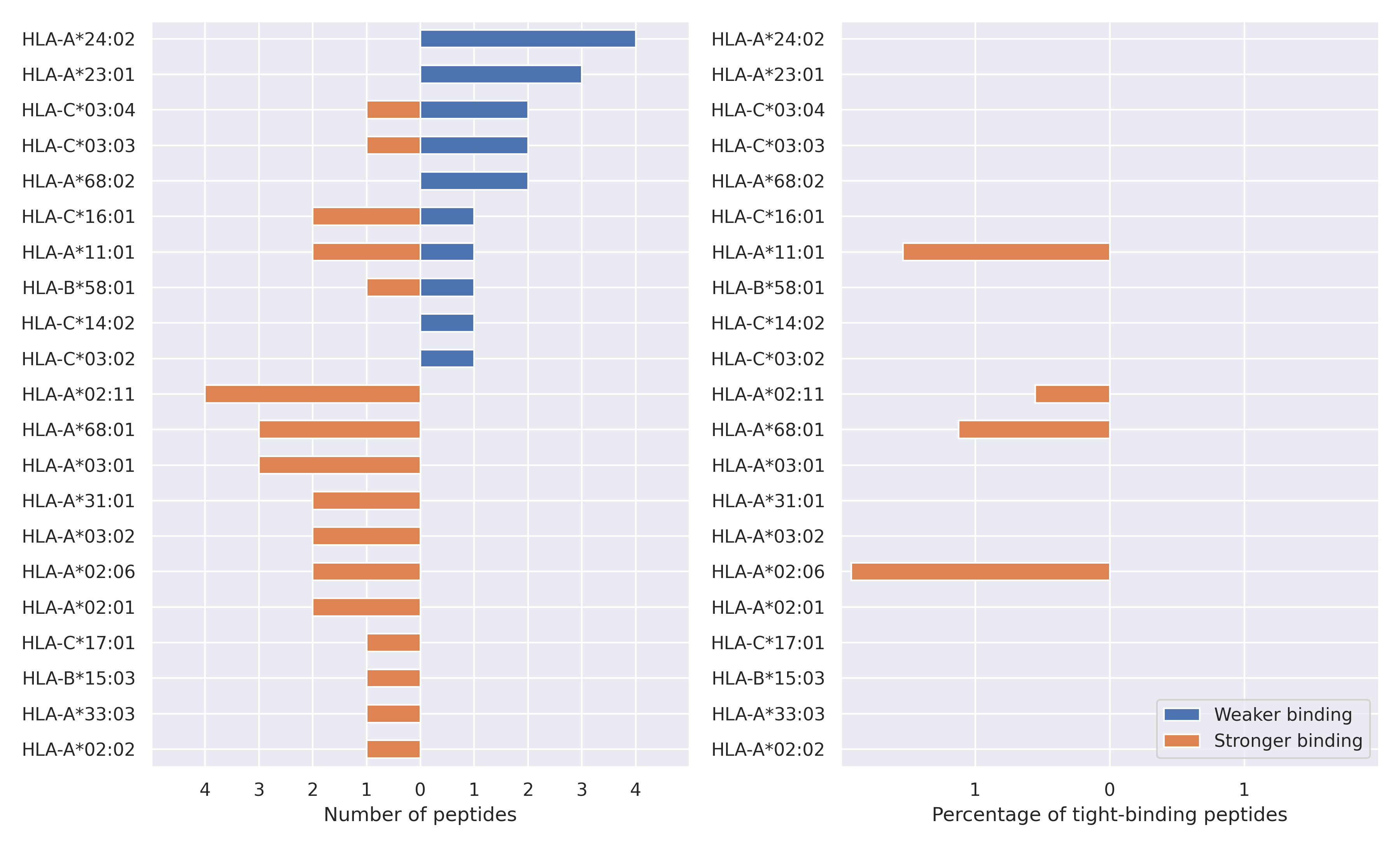
T24I
Reference APTKVTFGDDTVIEVQGYKSVNITFELDERIDKVLNEKCSAYTVELGTE
||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated APTKVTFGDDTVIEVQGYKSVNIIFELDERIDKVLNEKCSAYTVELGTE
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-B*58:01 | ITFELDERI | IIFELDERI | 270 | 4454 |
| HLA-B*15:03 | VQGYKSVNIT | VQGYKSVNII | 3702 | 395 |
| HLA-A*02:01 | ITFELDERI | IIFELDERI | 1637 | 223 |
| HLA-A*02:11 | ITFELDERI | IIFELDERI | 261 | 37 |
| HLA-A*02:02 | ITFELDERI | IIFELDERI | 428 | 65 |
| HLA-A*24:02 | VQGYKSVNITF | VQGYKSVNIIF | 194 | 811 |
| HLA-A*24:02 | IEVQGYKSVNITF | IEVQGYKSVNIIF | 366 | 1499 |
| HLA-A*68:02 | ITFELDERI | IIFELDERI | 253 | 1036 |
| HLA-A*68:01 | NITFELDER | NIIFELDER | 68 | 20 |
| HLA-A*33:03 | NITFELDER | NIIFELDER | 874 | 262 |
| HLA-A*23:01 | IEVQGYKSVNITF | IEVQGYKSVNIIF | 191 | 635 |
| HLA-C*03:03 | YKSVNITFEL | YKSVNIIFEL | 305 | 1007 |
| HLA-C*03:04 | YKSVNITFEL | YKSVNIIFEL | 305 | 1007 |
| HLA-A*23:01 | VIEVQGYKSVNITF | VIEVQGYKSVNIIF | 413 | 1337 |
| HLA-A*23:01 | VQGYKSVNITF | VQGYKSVNIIF | 111 | 353 |
| HLA-A*02:11 | KSVNITFEL | KSVNIIFEL | 169 | 55 |
| HLA-C*14:02 | GYKSVNITF | GYKSVNIIF | 53 | 160 |
| HLA-A*11:01 | ITFELDERIDK | IIFELDERIDK | 446 | 1259 |
| HLA-A*02:01 | KSVNITFEL | KSVNIIFEL | 863 | 307 |
| HLA-C*03:02 | YKSVNITFEL | YKSVNIIFEL | 299 | 784 |
| HLA-A*68:01 | VNITFELDER | VNIIFELDER | 1032 | 394 |
| HLA-A*02:06 | KSVNITFEL | KSVNIIFEL | 80 | 31 |
| HLA-C*16:01 | YKSVNITFEL | YKSVNIIFEL | 310 | 793 |
| HLA-A*68:02 | NITFELDERI | NIIFELDERI | 209 | 533 |
| HLA-A*24:02 | GYKSVNITF | GYKSVNIIF | 83 | 192 |
| HLA-A*02:11 | ITFELDERIDKV | IIFELDERIDKV | 397 | 175 |
| HLA-A*68:01 | SVNITFELDER | SVNIIFELDER | 405 | 180 |
| HLA-A*24:02 | QGYKSVNITF | QGYKSVNIIF | 208 | 455 |
| HLA-A*02:06 | ITFELDERI | IIFELDERI | 402 | 185 |
| HLA-A*02:11 | YKSVNITFEL | YKSVNIIFEL | 715 | 337 |
| HLA-C*03:03 | KSVNITFEL | KSVNIIFEL | 115 | 233 |
| HLA-C*03:04 | KSVNITFEL | KSVNIIFEL | 115 | 233 |
G489S
Reference DAPYIVGDVVQEGVLTAVVIPTKKAGGTTEMLAKALRKVPTDNYITTYPGQ
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated DAPYIVGDVVQEGVLTAVVIPTKKASGTTEMLAKALRKVPTDNYITTYPGQ
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-A*11:01 | AGGTTEMLAK | ASGTTEMLAK | 1254 | 37 |
| HLA-A*03:01 | AGGTTEMLAK | ASGTTEMLAK | 3665 | 277 |
| HLA-A*03:02 | AGGTTEMLAK | ASGTTEMLAK | 2689 | 228 |
| HLA-C*03:03 | KAGGTTEML | KASGTTEML | 2418 | 212 |
| HLA-C*03:04 | KAGGTTEML | KASGTTEML | 2418 | 212 |
| HLA-C*16:01 | KAGGTTEML | KASGTTEML | 1012 | 116 |
| HLA-C*17:01 | KAGGTTEML | KASGTTEML | 1802 | 287 |
| HLA-A*31:01 | AGGTTEMLAKALR | ASGTTEMLAKALR | 787 | 133 |
| HLA-B*58:01 | KAGGTTEML | KASGTTEML | 2117 | 440 |
| HLA-C*16:01 | KAGGTTEM | KASGTTEM | 863 | 191 |
| HLA-A*03:02 | GGTTEMLAKALRK | SGTTEMLAKALRK | 1693 | 453 |
| HLA-A*11:01 | KAGGTTEMLAK | KASGTTEMLAK | 589 | 203 |
| HLA-A*03:01 | KAGGTTEMLAK | KASGTTEMLAK | 1236 | 449 |
| HLA-A*03:01 | GGTTEMLAKALRK | SGTTEMLAKALRK | 467 | 181 |
| HLA-A*31:01 | GGTTEMLAKALR | SGTTEMLAKALR | 677 | 335 |
NSP4 protein
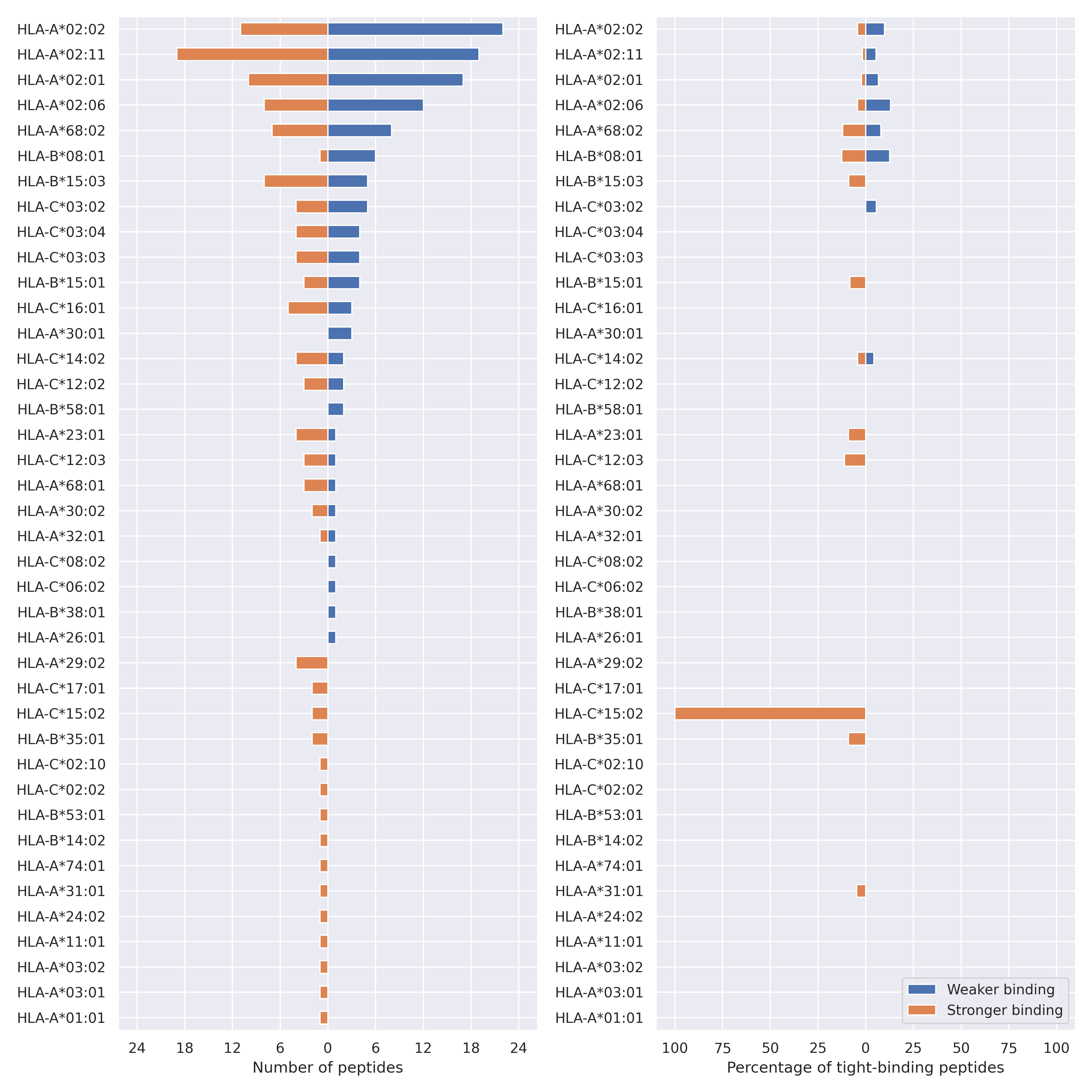
L264F
Reference GRWVLNNDYYRSLPGVFCGVDAVNLLTNMFTPLIQPIGALDISASIVAGGI
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated GRWVLNNDYYRSLPGVFCGVDAVNLFTNMFTPLIQPIGALDISASIVAGGI
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-A*02:11 | LLTNMFTPL | LFTNMFTPL | 5 | 2700 |
| HLA-A*02:01 | LLTNMFTPL | LFTNMFTPL | 16 | 6477 |
| HLA-A*02:02 | LLTNMFTPL | LFTNMFTPL | 5 | 1518 |
| HLA-A*02:06 | LLTNMFTPL | LFTNMFTPL | 28 | 3590 |
| HLA-A*02:11 | LLTNMFTPLI | LFTNMFTPLI | 17 | 1231 |
| HLA-A*02:01 | LLTNMFTPLI | LFTNMFTPLI | 55 | 3749 |
| HLA-C*14:02 | LLTNMFTPL | LFTNMFTPL | 453 | 7 |
| HLA-A*02:02 | LLTNMFTPLI | LFTNMFTPLI | 22 | 967 |
| HLA-B*15:01 | LLTNMFTPL | LFTNMFTPL | 228 | 5949 |
| HLA-C*03:03 | LTNMFTPL | FTNMFTPL | 1113 | 61 |
| HLA-C*03:04 | LTNMFTPL | FTNMFTPL | 1113 | 61 |
| HLA-A*23:01 | VFCGVDAVNLL | VFCGVDAVNLF | 2671 | 151 |
| HLA-A*02:02 | LLTNMFTPLIQPI | LFTNMFTPLIQPI | 294 | 4718 |
| HLA-A*02:02 | GVFCGVDAVNLL | GVFCGVDAVNLF | 439 | 6440 |
| HLA-C*14:02 | NLLTNMFTPL | NLFTNMFTPL | 4861 | 337 |
| HLA-B*15:03 | CGVDAVNLL | CGVDAVNLF | 6087 | 490 |
| HLA-A*02:11 | LLTNMFTPLIQPI | LFTNMFTPLIQPI | 297 | 3332 |
| HLA-C*03:02 | LTNMFTPL | FTNMFTPL | 829 | 77 |
| HLA-C*03:03 | LTNMFTPLI | FTNMFTPLI | 2207 | 207 |
| HLA-C*03:04 | LTNMFTPLI | FTNMFTPLI | 2207 | 207 |
| HLA-A*02:01 | LTNMFTPLI | FTNMFTPLI | 1097 | 103 |
| HLA-A*02:11 | LTNMFTPLI | FTNMFTPLI | 136 | 13 |
| HLA-C*12:03 | LTNMFTPLI | FTNMFTPLI | 430 | 42 |
| HLA-A*02:06 | LLTNMFTPLI | LFTNMFTPLI | 136 | 1345 |
| HLA-A*02:06 | LTNMFTPLI | FTNMFTPLI | 223 | 23 |
| HLA-C*12:02 | LTNMFTPLI | FTNMFTPLI | 1404 | 157 |
| HLA-A*02:02 | LTNMFTPLI | FTNMFTPLI | 364 | 41 |
| HLA-A*68:02 | LTNMFTPLIQPI | FTNMFTPLIQPI | 1086 | 129 |
| HLA-C*12:03 | LTNMFTPL | FTNMFTPL | 2677 | 324 |
| HLA-C*03:02 | LTNMFTPLI | FTNMFTPLI | 716 | 87 |
| HLA-C*02:02 | LTNMFTPLI | FTNMFTPLI | 1268 | 158 |
| HLA-C*02:10 | LTNMFTPLI | FTNMFTPLI | 1268 | 158 |
| HLA-C*17:01 | LTNMFTPLI | FTNMFTPLI | 599 | 86 |
| HLA-A*68:02 | LTNMFTPLI | FTNMFTPLI | 82 | 12 |
| HLA-C*03:03 | LLTNMFTPL | LFTNMFTPL | 253 | 1728 |
| HLA-C*03:04 | LLTNMFTPL | LFTNMFTPL | 253 | 1728 |
| HLA-C*12:02 | LTNMFTPL | FTNMFTPL | 3002 | 450 |
| HLA-B*15:03 | LLTNMFTPL | LFTNMFTPL | 108 | 704 |
| HLA-A*68:02 | NLLTNMFTPL | NLFTNMFTPL | 2741 | 480 |
| HLA-C*14:02 | LTNMFTPL | FTNMFTPL | 2589 | 467 |
| HLA-A*02:06 | LTNMFTPLIQPI | FTNMFTPLIQPI | 2171 | 394 |
| HLA-C*16:01 | LTNMFTPL | FTNMFTPL | 293 | 61 |
| HLA-C*16:01 | LTNMFTPLI | FTNMFTPLI | 267 | 56 |
| HLA-A*30:01 | LTNMFTPLI | FTNMFTPLI | 144 | 648 |
| HLA-A*02:02 | AVNLLTNMFTPL | AVNLFTNMFTPL | 249 | 948 |
| HLA-B*08:01 | LLTNMFTPL | LFTNMFTPL | 271 | 983 |
| HLA-A*02:11 | LTNMFTPLIQPI | FTNMFTPLIQPI | 1413 | 409 |
| HLA-C*03:02 | LLTNMFTPL | LFTNMFTPL | 307 | 951 |
| HLA-A*02:02 | NLLTNMFTPLI | NLFTNMFTPLI | 658 | 224 |
| HLA-A*02:02 | NLLTNMFTPL | NLFTNMFTPL | 58 | 21 |
| HLA-C*15:02 | LTNMFTPLI | FTNMFTPLI | 67 | 25 |
| HLA-A*02:11 | AVNLLTNMFTPL | AVNLFTNMFTPL | 390 | 1009 |
| HLA-A*02:02 | VNLLTNMFTPL | VNLFTNMFTPL | 375 | 919 |
| HLA-A*02:11 | NLLTNMFTP | NLFTNMFTP | 1166 | 476 |
| HLA-B*15:03 | AVNLLTNMF | AVNLFTNMF | 390 | 172 |
| HLA-A*02:11 | NLLTNMFTPLIQPI | NLFTNMFTPLIQPI | 274 | 129 |
T327I
Reference MRFRRAFGEYSHVVAFNTLLFLMSFTVLCLTPVYSFLPGVYSVIYLYLTFY
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated MRFRRAFGEYSHVVAFNTLLFLMSFIVLCLTPVYSFLPGVYSVIYLYLTFY
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-A*02:11 | NTLLFLMSFT | NTLLFLMSFI | 1059 | 51 |
| HLA-A*68:02 | NTLLFLMSFT | NTLLFLMSFI | 741 | 41 |
| HLA-A*02:11 | FNTLLFLMSFT | FNTLLFLMSFI | 1509 | 110 |
| HLA-A*02:11 | TLLFLMSFT | TLLFLMSFI | 82 | 6 |
| HLA-A*68:02 | TLLFLMSFT | TLLFLMSFI | 3858 | 286 |
| HLA-A*68:02 | FTVLCLTPVYSFL | FIVLCLTPVYSFL | 129 | 1580 |
| HLA-A*02:11 | AFNTLLFLMSFT | AFNTLLFLMSFI | 4598 | 420 |
| HLA-A*68:02 | FTVLCLTPV | FIVLCLTPV | 8 | 83 |
| HLA-A*68:02 | SFTVLCLTPV | SFIVLCLTPV | 63 | 597 |
| HLA-A*02:01 | TLLFLMSFT | TLLFLMSFI | 239 | 26 |
| HLA-A*02:01 | NTLLFLMSFT | NTLLFLMSFI | 1593 | 179 |
| HLA-A*02:02 | TLLFLMSFT | TLLFLMSFI | 235 | 28 |
| HLA-B*15:01 | LLFLMSFTVL | LLFLMSFIVL | 229 | 1839 |
| HLA-A*02:11 | LFLMSFTV | LFLMSFIV | 279 | 2222 |
| HLA-A*02:02 | NTLLFLMSFT | NTLLFLMSFI | 1462 | 186 |
| HLA-A*02:11 | LLFLMSFT | LLFLMSFI | 2935 | 377 |
| HLA-A*32:01 | LLFLMSFTV | LLFLMSFIV | 369 | 2631 |
| HLA-A*02:06 | LLFLMSFTV | LLFLMSFIV | 34 | 231 |
| HLA-C*12:03 | FTVLCLTPV | FIVLCLTPV | 161 | 1041 |
| HLA-C*14:02 | LFLMSFTVL | LFLMSFIVL | 18 | 112 |
| HLA-A*02:11 | AFNTLLFLMSFTV | AFNTLLFLMSFIV | 74 | 448 |
| HLA-C*16:01 | FTVLCLTPVY | FIVLCLTPVY | 395 | 2359 |
| HLA-A*02:06 | TLLFLMSFT | TLLFLMSFI | 397 | 68 |
| HLA-A*02:01 | LLFLMSFTV | LLFLMSFIV | 9 | 50 |
| HLA-A*02:11 | VAFNTLLFLMSFTV | VAFNTLLFLMSFIV | 225 | 1206 |
| HLA-A*02:01 | AFNTLLFLMSFTV | AFNTLLFLMSFIV | 357 | 1842 |
| HLA-A*68:02 | NTLLFLMSFTV | NTLLFLMSFIV | 106 | 539 |
| HLA-A*02:11 | NTLLFLMSFTV | NTLLFLMSFIV | 59 | 300 |
| HLA-A*02:01 | NTLLFLMSFTV | NTLLFLMSFIV | 212 | 1074 |
| HLA-C*03:03 | FTVLCLTPV | FIVLCLTPV | 302 | 1526 |
| HLA-C*03:04 | FTVLCLTPV | FIVLCLTPV | 302 | 1526 |
| HLA-B*15:03 | LLFLMSFTVL | LLFLMSFIVL | 318 | 1563 |
| HLA-A*02:02 | LLFLMSFTV | LLFLMSFIV | 11 | 53 |
| HLA-A*02:06 | NTLLFLMSFT | NTLLFLMSFI | 945 | 197 |
| HLA-A*02:11 | SFTVLCLTPV | SFIVLCLTPV | 250 | 53 |
| HLA-A*02:11 | FNTLLFLMSFTV | FNTLLFLMSFIV | 23 | 108 |
| HLA-A*02:01 | FNTLLFLMSFTV | FNTLLFLMSFIV | 131 | 602 |
| HLA-C*03:02 | FTVLCLTPV | FIVLCLTPV | 198 | 902 |
| HLA-C*14:02 | FLMSFTVL | FLMSFIVL | 475 | 2138 |
| HLA-C*12:02 | FTVLCLTPV | FIVLCLTPV | 218 | 961 |
| HLA-C*03:03 | FLMSFTVL | FLMSFIVL | 309 | 1359 |
| HLA-C*03:04 | FLMSFTVL | FLMSFIVL | 309 | 1359 |
| HLA-A*26:01 | FTVLCLTPVY | FIVLCLTPVY | 176 | 760 |
| HLA-A*02:01 | SFTVLCLTPV | SFIVLCLTPV | 470 | 111 |
| HLA-B*15:01 | FLMSFTVLCL | FLMSFIVLCL | 455 | 1914 |
| HLA-B*15:03 | LFLMSFTVL | LFLMSFIVL | 353 | 1462 |
| HLA-A*68:01 | FTVLCLTPVY | FIVLCLTPVY | 485 | 1972 |
| HLA-A*02:02 | FNTLLFLMSFTV | FNTLLFLMSFIV | 186 | 704 |
| HLA-B*15:01 | LMSFTVLCL | LMSFIVLCL | 110 | 408 |
| HLA-C*03:02 | FLMSFTVL | FLMSFIVL | 352 | 1285 |
| HLA-A*02:02 | NTLLFLMSFTV | NTLLFLMSFIV | 424 | 1515 |
| HLA-A*02:11 | LLFLMSFTV | LLFLMSFIV | 2 | 7 |
| HLA-C*03:02 | FTVLCLTPVY | FIVLCLTPVY | 50 | 175 |
| HLA-B*15:03 | LMSFTVLCL | LMSFIVLCL | 54 | 186 |
| HLA-C*12:02 | FTVLCLTPVY | FIVLCLTPVY | 331 | 1137 |
| HLA-A*02:02 | LLFLMSFTVL | LLFLMSFIVL | 42 | 144 |
| HLA-A*68:02 | MSFTVLCLTPV | MSFIVLCLTPV | 34 | 114 |
| HLA-C*03:02 | LMSFTVLCL | LMSFIVLCL | 467 | 1520 |
| HLA-A*02:01 | LLFLMSFTVL | LLFLMSFIVL | 74 | 240 |
| HLA-A*02:11 | LLFLMSFTVL | LLFLMSFIVL | 25 | 80 |
| HLA-A*02:02 | SFTVLCLTPV | SFIVLCLTPV | 261 | 84 |
| HLA-A*02:02 | LFLMSFTVLCL | LFLMSFIVLCL | 73 | 216 |
| HLA-A*02:06 | LLFLMSFTVL | LLFLMSFIVL | 331 | 973 |
| HLA-C*16:01 | MSFTVLCL | MSFIVLCL | 171 | 496 |
| HLA-A*68:02 | FLMSFTVLCLTPV | FLMSFIVLCLTPV | 331 | 927 |
| HLA-A*68:02 | MSFTVLCLT | MSFIVLCLT | 68 | 186 |
| HLA-A*02:06 | MSFTVLCLT | MSFIVLCLT | 239 | 650 |
| HLA-A*02:11 | TVLCLTPVYSFL | IVLCLTPVYSFL | 533 | 199 |
| HLA-B*15:03 | FLMSFTVL | FLMSFIVL | 371 | 993 |
| HLA-A*02:02 | FLMSFTVLCL | FLMSFIVLCL | 3 | 8 |
| HLA-A*02:02 | LLFLMSFTVLCL | LLFLMSFIVLCL | 134 | 356 |
| HLA-A*02:01 | LLFLMSFTVLCL | LLFLMSFIVLCL | 237 | 602 |
| HLA-A*02:01 | LFLMSFTVLCL | LFLMSFIVLCL | 73 | 185 |
| HLA-A*02:06 | FLMSFTVLCL | FLMSFIVLCL | 15 | 38 |
| HLA-C*03:02 | TVLCLTPVY | IVLCLTPVY | 136 | 54 |
| HLA-A*23:01 | LFLMSFTVL | LFLMSFIVL | 411 | 1026 |
| HLA-A*02:06 | TLLFLMSFTV | TLLFLMSFIV | 47 | 116 |
| HLA-B*08:01 | FLMSFTVL | FLMSFIVL | 118 | 291 |
| HLA-A*02:06 | NTLLFLMSFTV | NTLLFLMSFIV | 373 | 906 |
| HLA-A*02:01 | TLLFLMSFTV | TLLFLMSFIV | 14 | 33 |
| HLA-A*02:06 | VAFNTLLFLMSFTV | VAFNTLLFLMSFIV | 479 | 1101 |
| HLA-A*02:06 | MSFTVLCLTPV | MSFIVLCLTPV | 48 | 110 |
| HLA-B*58:01 | MSFTVLCLT | MSFIVLCLT | 412 | 943 |
| HLA-A*02:01 | FLMSFTVLCL | FLMSFIVLCL | 4 | 9 |
| HLA-A*02:02 | FLMSFTVLCLTP | FLMSFIVLCLTP | 194 | 430 |
| HLA-A*02:11 | TLLFLMSFTV | TLLFLMSFIV | 5 | 11 |
| HLA-A*02:02 | FTVLCLTPV | FIVLCLTPV | 23 | 11 |
| HLA-A*02:01 | FTVLCLTPV | FIVLCLTPV | 25 | 12 |
| HLA-A*30:02 | FTVLCLTPVY | FIVLCLTPVY | 327 | 671 |
| HLA-A*02:02 | TVLCLTPVYSFL | IVLCLTPVYSFL | 176 | 86 |
| HLA-B*58:01 | MSFTVLCLTP | MSFIVLCLTP | 497 | 1015 |
| HLA-A*02:11 | FTVLCLTPV | FIVLCLTPV | 12 | 6 |
L438F
Reference EEAALCTFLLNKEMYLKLRSDVLLPLTQYNRYLALYNKYKYFSGAMDTTSY
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated EEAALCTFLLNKEMYLKLRSDVLLPFTQYNRYLALYNKYKYFSGAMDTTSY
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-A*02:11 | YLKLRSDVLLPL | YLKLRSDVLLPF | 5 | 567 |
| HLA-A*02:02 | MYLKLRSDVLLPL | MYLKLRSDVLLPF | 49 | 5402 |
| HLA-A*02:11 | KLRSDVLLPL | KLRSDVLLPF | 5 | 501 |
| HLA-A*02:01 | KLRSDVLLPL | KLRSDVLLPF | 10 | 977 |
| HLA-A*02:01 | YLKLRSDVLLPL | YLKLRSDVLLPF | 11 | 948 |
| HLA-A*02:11 | LKLRSDVLLPL | LKLRSDVLLPF | 93 | 7534 |
| HLA-A*02:11 | MYLKLRSDVLLPL | MYLKLRSDVLLPF | 82 | 6537 |
| HLA-A*02:02 | YLKLRSDVLLPL | YLKLRSDVLLPF | 6 | 468 |
| HLA-A*02:02 | LKLRSDVLLPL | LKLRSDVLLPF | 120 | 9124 |
| HLA-A*02:02 | KLRSDVLLPL | KLRSDVLLPF | 6 | 425 |
| HLA-A*02:01 | LKLRSDVLLPL | LKLRSDVLLPF | 161 | 9994 |
| HLA-A*02:01 | MYLKLRSDVLLPL | MYLKLRSDVLLPF | 161 | 8888 |
| HLA-A*02:02 | LPLTQYNRYL | LPFTQYNRYL | 241 | 12319 |
| HLA-A*02:06 | YLKLRSDVLLPL | YLKLRSDVLLPF | 35 | 1391 |
| HLA-A*02:01 | KLRSDVLLPLT | KLRSDVLLPFT | 144 | 5015 |
| HLA-A*02:02 | YLKLRSDVLLPLT | YLKLRSDVLLPFT | 82 | 2744 |
| HLA-A*02:02 | KLRSDVLLPLT | KLRSDVLLPFT | 183 | 5811 |
| HLA-A*02:02 | EMYLKLRSDVLLPL | EMYLKLRSDVLLPF | 322 | 10179 |
| HLA-A*02:06 | KLRSDVLLPL | KLRSDVLLPF | 29 | 851 |
| HLA-A*02:11 | EMYLKLRSDVLLPL | EMYLKLRSDVLLPF | 400 | 11514 |
| HLA-B*15:01 | KLRSDVLLPL | KLRSDVLLPF | 335 | 15 |
| HLA-B*15:01 | YLKLRSDVLLPL | YLKLRSDVLLPF | 1921 | 89 |
| HLA-A*02:11 | KLRSDVLLPLT | KLRSDVLLPFT | 114 | 2241 |
| HLA-B*15:03 | KLRSDVLLPL | KLRSDVLLPF | 740 | 38 |
| HLA-B*15:03 | LKLRSDVLLPL | LKLRSDVLLPF | 752 | 42 |
| HLA-C*03:03 | LTQYNRYLAL | FTQYNRYLAL | 922 | 58 |
| HLA-C*03:04 | LTQYNRYLAL | FTQYNRYLAL | 922 | 58 |
| HLA-A*02:01 | YLKLRSDVLLPLT | YLKLRSDVLLPFT | 177 | 2552 |
| HLA-B*15:03 | YLKLRSDVLLPL | YLKLRSDVLLPF | 2864 | 206 |
| HLA-B*15:03 | LRSDVLLPL | LRSDVLLPF | 149 | 11 |
| HLA-A*23:01 | MYLKLRSDVLLPL | MYLKLRSDVLLPF | 466 | 39 |
| HLA-B*15:03 | MYLKLRSDVLLPL | MYLKLRSDVLLPF | 3874 | 334 |
| HLA-A*02:11 | LTQYNRYLAL | FTQYNRYLAL | 1343 | 121 |
| HLA-A*02:11 | YLKLRSDVLLPLT | YLKLRSDVLLPFT | 115 | 1240 |
| HLA-B*15:03 | RSDVLLPL | RSDVLLPF | 4307 | 407 |
| HLA-C*12:03 | LTQYNRYLAL | FTQYNRYLAL | 779 | 75 |
| HLA-A*24:02 | MYLKLRSDVLLPL | MYLKLRSDVLLPF | 1229 | 123 |
| HLA-A*68:02 | LTQYNRYLA | FTQYNRYLA | 349 | 38 |
| HLA-A*68:02 | LTQYNRYLAL | FTQYNRYLAL | 1524 | 172 |
| HLA-A*02:11 | LTQYNRYLA | FTQYNRYLA | 2873 | 342 |
| HLA-C*03:02 | LTQYNRYLAL | FTQYNRYLAL | 539 | 66 |
| HLA-C*12:02 | LTQYNRYLAL | FTQYNRYLAL | 1553 | 198 |
| HLA-A*02:02 | LTQYNRYLAL | FTQYNRYLAL | 1070 | 154 |
| HLA-A*02:06 | LTQYNRYLAL | FTQYNRYLAL | 483 | 77 |
| HLA-A*68:01 | LTQYNRYLALYNK | FTQYNRYLALYNK | 793 | 128 |
| HLA-B*08:01 | EMYLKLRSDVLLPL | EMYLKLRSDVLLPF | 231 | 1402 |
| HLA-C*17:01 | LTQYNRYLAL | FTQYNRYLAL | 2488 | 410 |
| HLA-A*02:01 | LTQYNRYLAL | FTQYNRYLAL | 2931 | 490 |
| HLA-A*32:01 | KLRSDVLLPL | KLRSDVLLPF | 418 | 70 |
| HLA-B*08:01 | MYLKLRSDVLLPL | MYLKLRSDVLLPF | 151 | 847 |
| HLA-C*16:01 | LTQYNRYLAL | FTQYNRYLAL | 285 | 51 |
| HLA-C*16:01 | LTQYNRYL | FTQYNRYL | 663 | 140 |
| HLA-B*08:01 | YLKLRSDVLLPL | YLKLRSDVLLPF | 13 | 61 |
| HLA-C*14:02 | LTQYNRYLAL | FTQYNRYLAL | 1130 | 251 |
| HLA-A*30:01 | LTQYNRYLA | FTQYNRYLA | 171 | 719 |
| HLA-C*08:02 | RSDVLLPL | RSDVLLPF | 400 | 1636 |
| HLA-B*35:01 | LLPLTQYNRY | LLPFTQYNRY | 1778 | 445 |
| HLA-B*08:01 | YLKLRSDVLLPLT | YLKLRSDVLLPFT | 258 | 1028 |
| HLA-A*29:02 | LPLTQYNRY | LPFTQYNRY | 1848 | 480 |
| HLA-C*06:02 | LRSDVLLPL | LRSDVLLPF | 103 | 395 |
| HLA-B*35:01 | LPLTQYNRY | LPFTQYNRY | 66 | 18 |
| HLA-A*30:01 | KLRSDVLLPL | KLRSDVLLPF | 108 | 363 |
| HLA-C*15:02 | LTQYNRYLAL | FTQYNRYLAL | 541 | 163 |
| HLA-A*29:02 | LTQYNRYLALYNKY | FTQYNRYLALYNKY | 1609 | 497 |
| HLA-A*68:01 | LLPLTQYNR | LLPFTQYNR | 221 | 69 |
| HLA-A*11:01 | VLLPLTQYNR | VLLPFTQYNR | 881 | 285 |
| HLA-A*29:02 | LLPLTQYNRY | LLPFTQYNRY | 239 | 81 |
| HLA-A*29:02 | LTQYNRYLALY | FTQYNRYLALY | 212 | 76 |
| HLA-A*01:01 | LTQYNRYLALY | FTQYNRYLALY | 400 | 146 |
| HLA-B*08:01 | LTQYNRYLAL | FTQYNRYLAL | 126 | 46 |
| HLA-A*02:11 | LLPLTQYNRYL | LLPFTQYNRYL | 389 | 143 |
| HLA-A*30:02 | LLPLTQYNRY | LLPFTQYNRY | 547 | 211 |
| HLA-A*03:01 | VLLPLTQYNR | VLLPFTQYNR | 1129 | 439 |
| HLA-B*15:01 | LLPLTQYNRY | LLPFTQYNRY | 890 | 357 |
| HLA-B*14:02 | LTQYNRYLAL | FTQYNRYLAL | 712 | 291 |
| HLA-A*03:02 | VLLPLTQYNR | VLLPFTQYNR | 318 | 131 |
| HLA-A*68:01 | VLLPLTQYNR | VLLPFTQYNR | 604 | 250 |
| HLA-A*30:02 | RSDVLLPLTQYNRY | RSDVLLPFTQYNRY | 565 | 236 |
| HLA-B*53:01 | LPLTQYNRY | LPFTQYNRY | 497 | 208 |
| HLA-A*02:11 | VLLPLTQYNRYLA | VLLPFTQYNRYLA | 229 | 98 |
| HLA-A*02:01 | VLLPLTQYNRYLA | VLLPFTQYNRYLA | 545 | 239 |
| HLA-A*74:01 | VLLPLTQYNR | VLLPFTQYNR | 513 | 226 |
| HLA-A*02:11 | VLLPLTQYNRYLAL | VLLPFTQYNRYLAL | 139 | 65 |
| HLA-B*38:01 | LRSDVLLPL | LRSDVLLPF | 499 | 1065 |
| HLA-A*02:02 | VLLPLTQYNRYLA | VLLPFTQYNRYLA | 980 | 473 |
| HLA-A*02:11 | VLLPLTQYNRYL | VLLPFTQYNRYL | 43 | 21 |
| HLA-A*02:02 | LLPLTQYNRYL | LLPFTQYNRYL | 400 | 196 |
| HLA-A*31:01 | VLLPLTQYNR | VLLPFTQYNR | 80 | 40 |
| HLA-A*02:01 | VLLPLTQYNRYL | VLLPFTQYNRYL | 170 | 85 |
T492I
Reference ACCHLAKALNDFSNSGSDVLYQPPQTSITSAVLQ
||||||||||||||||||||||||| ||||||||
Mutated ACCHLAKALNDFSNSGSDVLYQPPQISITSAVLQ
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-C*16:01 | QTSITSAVL | QISITSAVL | 201 | 1829 |
| HLA-A*68:02 | QTSITSAVL | QISITSAVL | 206 | 1702 |
| HLA-A*02:06 | QPPQTSITSAV | QPPQISITSAV | 3097 | 393 |
| HLA-A*02:06 | LYQPPQTSITSAV | LYQPPQISITSAV | 1485 | 214 |
| HLA-A*02:06 | YQPPQTSITSAV | YQPPQISITSAV | 156 | 26 |
| HLA-A*02:01 | YQPPQTSITSAV | YQPPQISITSAV | 1613 | 341 |
| HLA-A*02:11 | YQPPQTSITSAV | YQPPQISITSAV | 498 | 120 |
| HLA-C*03:03 | TSITSAVL | ISITSAVL | 532 | 198 |
| HLA-C*03:04 | TSITSAVL | ISITSAVL | 532 | 198 |
| HLA-C*03:03 | QTSITSAVL | QISITSAVL | 311 | 831 |
| HLA-C*03:04 | QTSITSAVL | QISITSAVL | 311 | 831 |
| HLA-C*16:01 | TSITSAVL | ISITSAVL | 381 | 156 |
| HLA-A*02:01 | VLYQPPQTSI | VLYQPPQISI | 1084 | 474 |
| HLA-A*23:01 | VLYQPPQTSI | VLYQPPQISI | 979 | 462 |
| HLA-A*23:01 | LYQPPQTSI | LYQPPQISI | 278 | 132 |
NSP5 protein
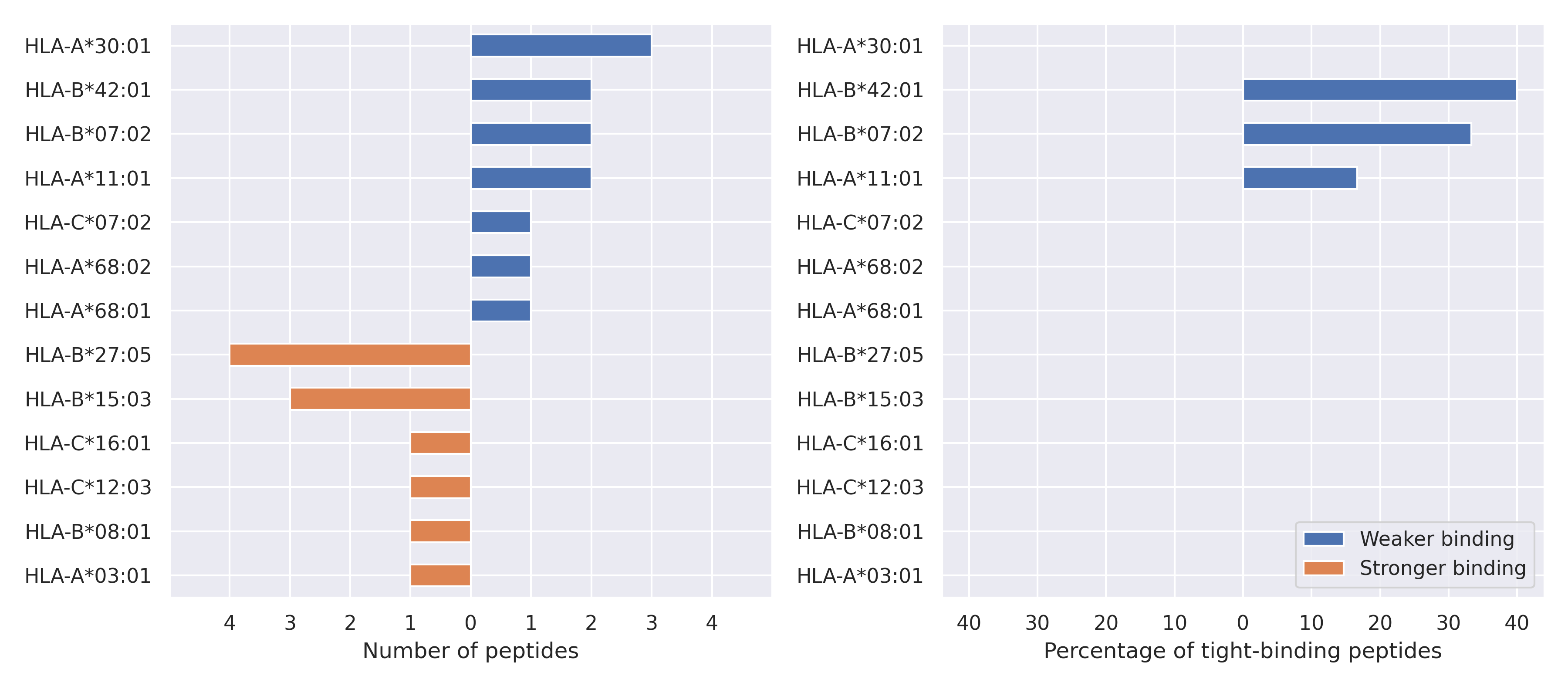
P132H
Reference QPGQTFSVLACYNGSPSGVYQCAMRPNFTIKGSFLNGSCGSVGFNIDYDCV
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated QPGQTFSVLACYNGSPSGVYQCAMRHNFTIKGSFLNGSCGSVGFNIDYDCV
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-B*07:02 | RPNFTIKGSF | RHNFTIKGSF | 14 | 15756 |
| HLA-B*07:02 | RPNFTIKGSFL | RHNFTIKGSFL | 24 | 18650 |
| HLA-B*42:01 | RPNFTIKGSF | RHNFTIKGSF | 31 | 15129 |
| HLA-B*42:01 | RPNFTIKGSFL | RHNFTIKGSFL | 39 | 17143 |
| HLA-B*15:03 | PNFTIKGSF | HNFTIKGSF | 7728 | 117 |
| HLA-A*03:01 | GVYQCAMRP | GVYQCAMRH | 20799 | 417 |
| HLA-B*08:01 | CAMRPNFTI | CAMRHNFTI | 3487 | 242 |
| HLA-B*27:05 | MRPNFTIKG | MRHNFTIKG | 4762 | 366 |
| HLA-B*27:05 | MRPNFTIKGSFL | MRHNFTIKGSFL | 827 | 86 |
| HLA-B*15:03 | RPNFTIKGSF | RHNFTIKGSF | 432 | 54 |
| HLA-B*27:05 | MRPNFTIK | MRHNFTIK | 2132 | 276 |
| HLA-B*15:03 | MRPNFTIKGSF | MRHNFTIKGSF | 397 | 96 |
| HLA-C*07:02 | MRPNFTIKGSF | MRHNFTIKGSF | 361 | 1054 |
| HLA-B*27:05 | MRPNFTIKGSF | MRHNFTIKGSF | 218 | 78 |
| HLA-A*30:01 | QCAMRPNFTIK | QCAMRHNFTIK | 199 | 513 |
| HLA-A*68:02 | CAMRPNFTI | CAMRHNFTI | 199 | 493 |
| HLA-C*16:01 | CAMRPNFTI | CAMRHNFTI | 125 | 52 |
| HLA-A*30:01 | AMRPNFTIKG | AMRHNFTIKG | 123 | 279 |
| HLA-C*12:03 | CAMRPNFTI | CAMRHNFTI | 317 | 143 |
| HLA-A*30:01 | VYQCAMRPNFTIK | VYQCAMRHNFTIK | 316 | 683 |
| HLA-A*11:01 | AMRPNFTIK | AMRHNFTIK | 175 | 376 |
| HLA-A*11:01 | CAMRPNFTIK | CAMRHNFTIK | 42 | 88 |
| HLA-A*68:01 | CAMRPNFTIK | CAMRHNFTIK | 127 | 254 |
NSP6 protein
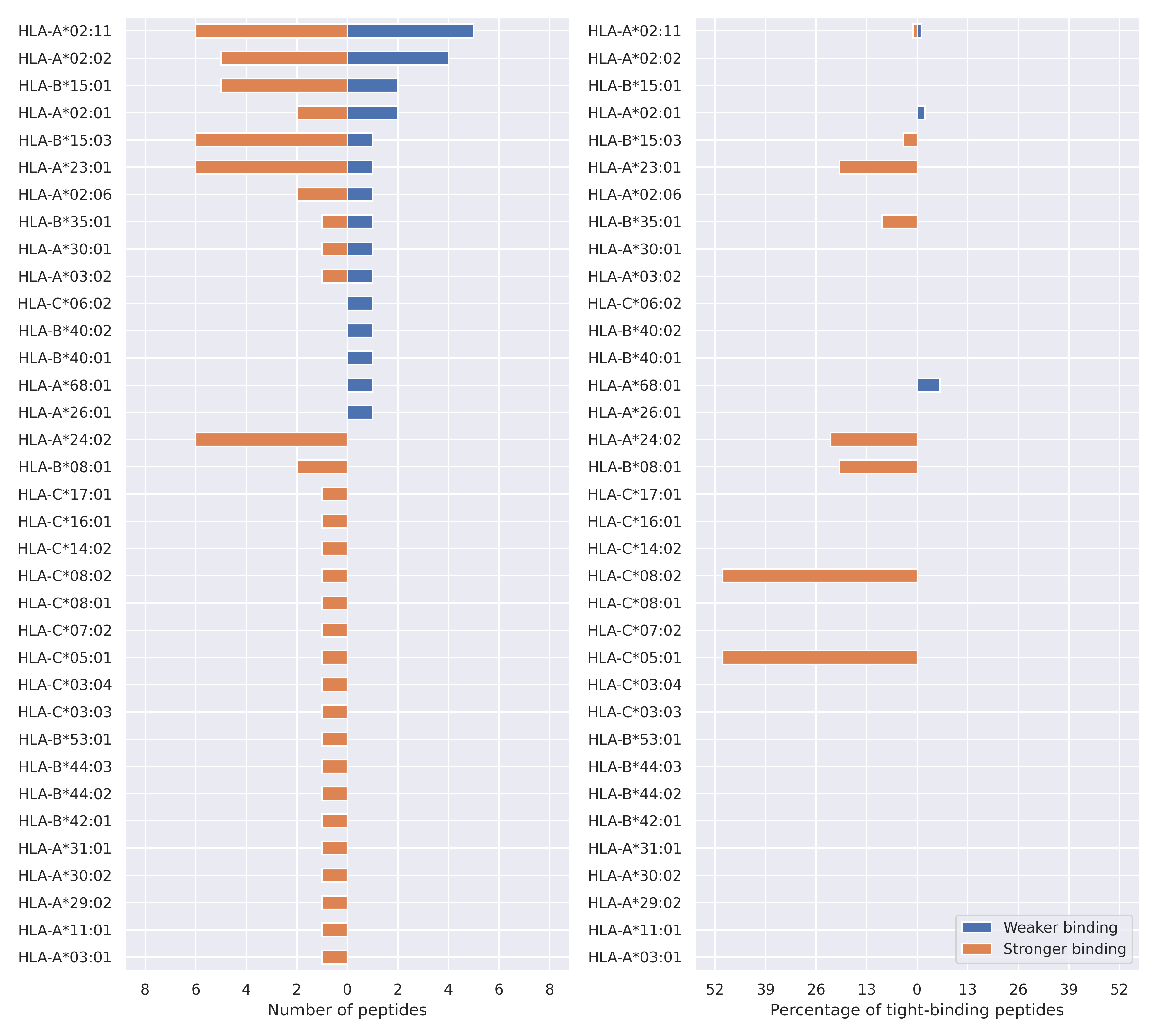
106-108 SGF->TSL
Reference FNMVYMPASWVMRIMTWLDMVDTSLSGFKLKDCVMYASAVVLLILMTARTVYD
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated FNMVYMPASWVMRIMTWLDMVDTSLTSLKLKDCVMYASAVVLLILMTARTVYD
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-A*02:11 | MVDTSLSGF | MVDTSLTSL | 4111 | 77 |
| HLA-B*15:01 | GFKLKDCVM | SLKLKDCVM | 9405 | 197 |
| HLA-B*15:01 | GFKLKDCVMY | SLKLKDCVMY | 7051 | 158 |
| HLA-B*08:01 | GFKLKDCVM | SLKLKDCVM | 1477 | 36 |
| HLA-A*02:11 | WLDMVDTSLSGF | WLDMVDTSLTSL | 6135 | 178 |
| HLA-A*02:02 | WLDMVDTSLSGF | WLDMVDTSLTSL | 4418 | 183 |
| HLA-C*03:03 | MVDTSLSGF | MVDTSLTSL | 1448 | 65 |
| HLA-C*03:04 | MVDTSLSGF | MVDTSLTSL | 1448 | 65 |
| HLA-A*02:02 | MVDTSLSGF | MVDTSLTSL | 4757 | 246 |
| HLA-C*08:01 | MVDTSLSGF | MVDTSLTSL | 3054 | 162 |
| HLA-C*17:01 | MVDTSLSGF | MVDTSLTSL | 1293 | 83 |
| HLA-A*02:06 | MVDTSLSGF | MVDTSLTSL | 1359 | 89 |
| HLA-B*15:03 | GFKLKDCVM | SLKLKDCVM | 4465 | 297 |
| HLA-A*26:01 | DMVDTSLSGF | DMVDTSLTSL | 417 | 5554 |
| HLA-B*15:01 | DMVDTSLSGF | DMVDTSLTSL | 362 | 4805 |
| HLA-A*23:01 | TWLDMVDTSLSGF | TWLDMVDTSLTSL | 387 | 3346 |
| HLA-C*08:02 | MVDTSLSGF | MVDTSLTSL | 159 | 19 |
| HLA-A*02:06 | FKLKDCVMYA | LKLKDCVMYA | 90 | 586 |
| HLA-A*02:01 | FKLKDCVMYA | LKLKDCVMYA | 27 | 165 |
| HLA-C*16:01 | MVDTSLSGF | MVDTSLTSL | 1460 | 255 |
| HLA-A*02:11 | SLSGFKLKDCV | SLTSLKLKDCV | 122 | 599 |
| HLA-B*15:03 | MVDTSLSGF | MVDTSLTSL | 276 | 1322 |
| HLA-A*02:11 | FKLKDCVMYA | LKLKDCVMYA | 12 | 54 |
| HLA-A*02:11 | SLSGFKLKDCVMYA | SLTSLKLKDCVMYA | 64 | 259 |
| HLA-A*02:02 | SLSGFKLKDCV | SLTSLKLKDCV | 408 | 1490 |
| HLA-A*02:02 | SLSGFKLKDCVMYA | SLTSLKLKDCVMYA | 55 | 198 |
| HLA-A*02:02 | FKLKDCVMYA | LKLKDCVMYA | 8 | 28 |
| HLA-B*35:01 | MVDTSLSGF | MVDTSLTSL | 490 | 1594 |
| HLA-B*08:01 | SGFKLKDCVM | TSLKLKDCVM | 920 | 289 |
| HLA-B*15:03 | GFKLKDCVMY | SLKLKDCVMY | 1062 | 349 |
| HLA-A*02:01 | SLSGFKLKDCVMYA | SLTSLKLKDCVMYA | 203 | 611 |
| HLA-C*05:01 | MVDTSLSGF | MVDTSLTSL | 64 | 23 |
| HLA-A*02:11 | FKLKDCVMYASAVV | LKLKDCVMYASAVV | 481 | 1324 |
| HLA-A*02:11 | SGFKLKDCVMYA | TSLKLKDCVMYA | 200 | 536 |
| HLA-A*68:01 | DTSLSGFKLK | DTSLTSLKLK | 33 | 80 |
L260F
Reference FRLTLGVYDYLVSTQEFRYMNSQGLLPPKNSIDAFKLNIKLLGVGGKPCIK
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated FRLTLGVYDYLVSTQEFRYMNSQGLFPPKNSIDAFKLNIKLLGVGGKPCIK
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-B*15:01 | LLPPKNSIDAF | LFPPKNSIDAF | 176 | 6276 |
| HLA-C*14:02 | LLPPKNSIDAF | LFPPKNSIDAF | 6426 | 271 |
| HLA-B*15:03 | YMNSQGLL | YMNSQGLF | 1229 | 53 |
| HLA-A*23:01 | FRYMNSQGLL | FRYMNSQGLF | 420 | 21 |
| HLA-A*29:02 | FRYMNSQGLL | FRYMNSQGLF | 2993 | 157 |
| HLA-A*24:02 | FRYMNSQGLL | FRYMNSQGLF | 397 | 21 |
| HLA-A*02:02 | YMNSQGLL | YMNSQGLF | 421 | 7283 |
| HLA-A*24:02 | TQEFRYMNSQGLL | TQEFRYMNSQGLF | 4225 | 247 |
| HLA-B*15:03 | TQEFRYMNSQGLL | TQEFRYMNSQGLF | 3079 | 191 |
| HLA-A*24:02 | QEFRYMNSQGLL | QEFRYMNSQGLF | 4406 | 281 |
| HLA-A*23:01 | TQEFRYMNSQGLL | TQEFRYMNSQGLF | 4468 | 286 |
| HLA-A*23:01 | QEFRYMNSQGLL | QEFRYMNSQGLF | 4157 | 271 |
| HLA-A*23:01 | EFRYMNSQGLL | EFRYMNSQGLF | 4327 | 294 |
| HLA-B*15:01 | TQEFRYMNSQGLL | TQEFRYMNSQGLF | 7065 | 483 |
| HLA-A*24:02 | EFRYMNSQGLL | EFRYMNSQGLF | 4788 | 337 |
| HLA-B*15:03 | FRYMNSQGLL | FRYMNSQGLF | 379 | 29 |
| HLA-B*15:01 | YMNSQGLL | YMNSQGLF | 3577 | 285 |
| HLA-B*15:03 | RYMNSQGLL | RYMNSQGLF | 714 | 62 |
| HLA-A*03:02 | LLPPKNSIDAFK | LFPPKNSIDAFK | 120 | 1314 |
| HLA-A*23:01 | RYMNSQGLL | RYMNSQGLF | 117 | 11 |
| HLA-B*40:01 | QEFRYMNSQGLL | QEFRYMNSQGLF | 252 | 2575 |
| HLA-B*44:03 | QEFRYMNSQGLL | QEFRYMNSQGLF | 1102 | 110 |
| HLA-A*24:02 | RYMNSQGLL | RYMNSQGLF | 95 | 10 |
| HLA-B*44:02 | QEFRYMNSQGLL | QEFRYMNSQGLF | 881 | 100 |
| HLA-A*24:02 | RYMNSQGLLP | RYMNSQGLFP | 3368 | 427 |
| HLA-B*35:01 | LPPKNSIDAF | FPPKNSIDAF | 316 | 42 |
| HLA-A*02:02 | GLLPPKNSIDAFKL | GLFPPKNSIDAFKL | 1648 | 221 |
| HLA-A*23:01 | RYMNSQGLLP | RYMNSQGLFP | 3140 | 451 |
| HLA-B*40:02 | QEFRYMNSQGLL | QEFRYMNSQGLF | 181 | 1214 |
| HLA-A*30:02 | RYMNSQGLL | RYMNSQGLF | 961 | 180 |
| HLA-B*53:01 | LPPKNSIDAF | FPPKNSIDAF | 1246 | 278 |
| HLA-A*30:01 | RYMNSQGLL | RYMNSQGLF | 320 | 1414 |
| HLA-A*03:01 | GLLPPKNSIDAFK | GLFPPKNSIDAFK | 251 | 62 |
| HLA-A*02:11 | YMNSQGLLPP | YMNSQGLFPP | 744 | 186 |
| HLA-A*02:02 | GLLPPKNSI | GLFPPKNSI | 634 | 175 |
| HLA-A*02:11 | GLLPPKNSIDAFKL | GLFPPKNSIDAFKL | 496 | 171 |
| HLA-A*02:01 | YMNSQGLLPP | YMNSQGLFPP | 1037 | 372 |
| HLA-B*15:01 | GLLPPKNSIDAF | GLFPPKNSIDAF | 867 | 313 |
| HLA-A*02:06 | YMNSQGLLPP | YMNSQGLFPP | 1140 | 419 |
| HLA-A*02:02 | YMNSQGLLPP | YMNSQGLFPP | 436 | 163 |
| HLA-C*06:02 | FRYMNSQGLL | FRYMNSQGLF | 63 | 160 |
| HLA-A*11:01 | GLLPPKNSIDAFK | GLFPPKNSIDAFK | 799 | 327 |
| HLA-A*02:01 | GLLPPKNSIDAFKL | GLFPPKNSIDAFKL | 953 | 424 |
| HLA-A*02:11 | YMNSQGLLPPKNSI | YMNSQGLFPPKNSI | 512 | 230 |
| HLA-A*03:02 | GLLPPKNSIDAFK | GLFPPKNSIDAFK | 191 | 88 |
| HLA-A*31:01 | NSQGLLPPK | NSQGLFPPK | 878 | 409 |
| HLA-B*42:01 | LPPKNSIDAF | FPPKNSIDAF | 322 | 151 |
| HLA-A*02:11 | GLLPPKNSI | GLFPPKNSI | 60 | 29 |
| HLA-A*30:01 | NSQGLLPPK | NSQGLFPPK | 188 | 93 |
| HLA-C*07:02 | FRYMNSQGLL | FRYMNSQGLF | 203 | 101 |
NSP12 protein
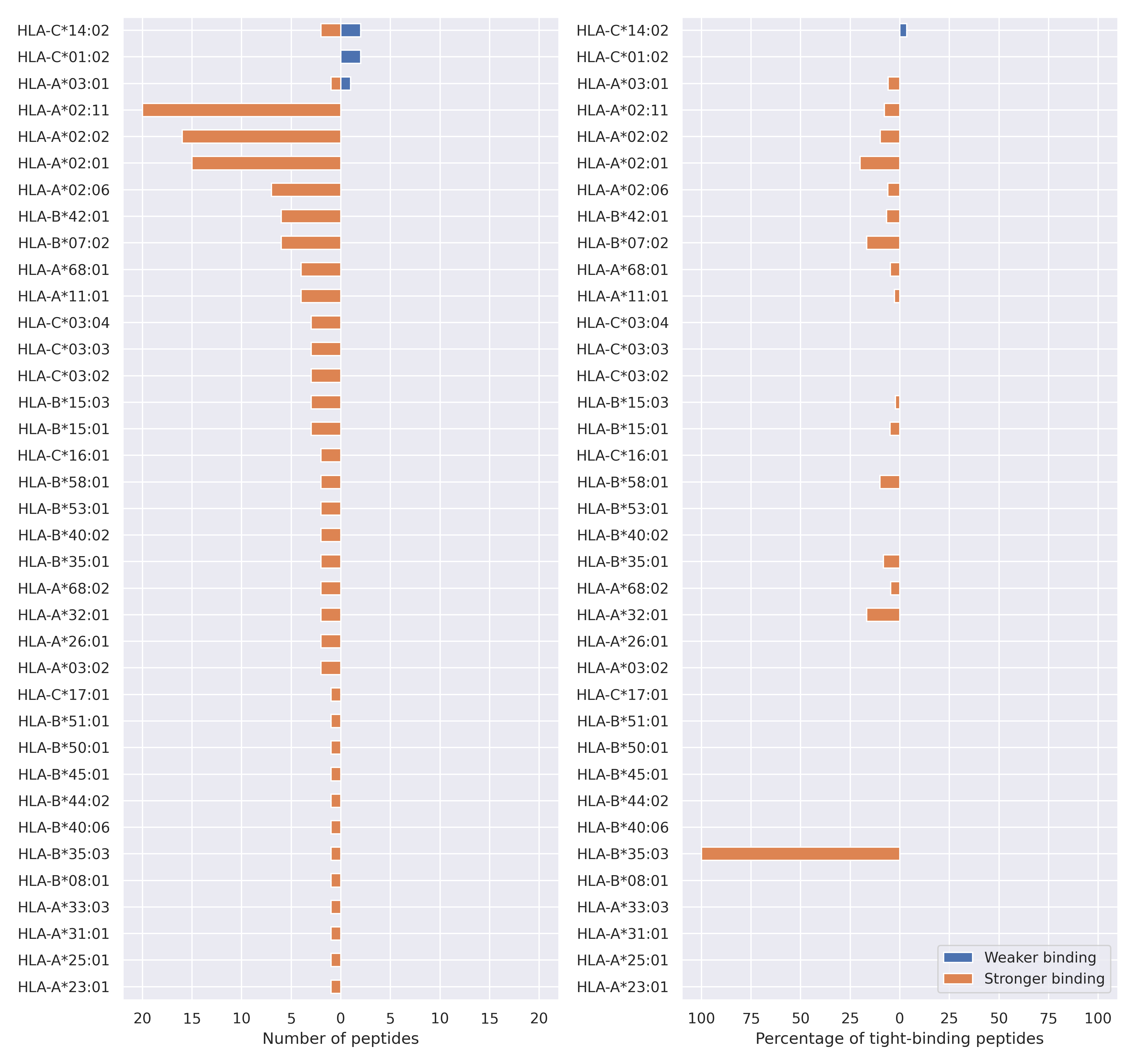
P323L
Reference CVNCLDDRCILHCANFNVLFSTVFPPTSFGPLVRKIFVDGVPFVVSTGYHF
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated CVNCLDDRCILHCANFNVLFSTVFPLTSFGPLVRKIFVDGVPFVVSTGYHF
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-A*02:01 | FPPTSFGPLV | FPLTSFGPLV | 8999 | 21 |
| HLA-A*02:02 | FPPTSFGPLV | FPLTSFGPLV | 7237 | 18 |
| HLA-A*02:11 | FPPTSFGPLV | FPLTSFGPLV | 3405 | 9 |
| HLA-A*32:01 | VLFSTVFPP | VLFSTVFPL | 5415 | 27 |
| HLA-A*02:11 | LFSTVFPP | LFSTVFPL | 6706 | 35 |
| HLA-A*02:11 | ANFNVLFSTVFPP | ANFNVLFSTVFPL | 1827 | 12 |
| HLA-A*02:11 | NFNVLFSTVFPP | NFNVLFSTVFPL | 1779 | 13 |
| HLA-A*02:11 | NVLFSTVFPP | NVLFSTVFPL | 739 | 6 |
| HLA-C*03:03 | VLFSTVFPP | VLFSTVFPL | 28468 | 265 |
| HLA-C*03:04 | VLFSTVFPP | VLFSTVFPL | 28468 | 265 |
| HLA-A*02:11 | FNVLFSTVFPP | FNVLFSTVFPL | 413 | 4 |
| HLA-C*14:02 | VLFSTVFPP | VLFSTVFPL | 21407 | 220 |
| HLA-A*02:11 | FNVLFSTVFPPT | FNVLFSTVFPLT | 2618 | 27 |
| HLA-A*02:01 | LFSTVFPP | LFSTVFPL | 7475 | 81 |
| HLA-C*14:02 | LFSTVFPP | LFSTVFPL | 11979 | 138 |
| HLA-B*15:03 | VLFSTVFPP | VLFSTVFPL | 3115 | 36 |
| HLA-A*02:11 | VLFSTVFPP | VLFSTVFPL | 78 | 1 |
| HLA-C*03:02 | VLFSTVFPP | VLFSTVFPL | 20533 | 273 |
| HLA-A*02:01 | FNVLFSTVFPP | FNVLFSTVFPL | 656 | 9 |
| HLA-A*02:01 | ANFNVLFSTVFPP | ANFNVLFSTVFPL | 2569 | 37 |
| HLA-A*02:11 | FNVLFSTVFPPTS | FNVLFSTVFPLTS | 9448 | 142 |
| HLA-A*02:02 | NVLFSTVFPP | NVLFSTVFPL | 1088 | 17 |
| HLA-A*02:11 | CANFNVLFSTVFPP | CANFNVLFSTVFPL | 6143 | 96 |
| HLA-A*02:02 | FNVLFSTVFPP | FNVLFSTVFPL | 1014 | 16 |
| HLA-A*68:02 | NVLFSTVFPP | NVLFSTVFPL | 2213 | 37 |
| HLA-A*02:02 | ANFNVLFSTVFPP | ANFNVLFSTVFPL | 3126 | 53 |
| HLA-A*02:11 | PPTSFGPLV | PLTSFGPLV | 28329 | 483 |
| HLA-C*16:01 | VLFSTVFPP | VLFSTVFPL | 24733 | 432 |
| HLA-C*17:01 | VLFSTVFPP | VLFSTVFPL | 25087 | 439 |
| HLA-A*02:01 | FNVLFSTVFPPT | FNVLFSTVFPLT | 3765 | 66 |
| HLA-A*02:02 | LFSTVFPP | LFSTVFPL | 9311 | 167 |
| HLA-A*02:01 | NVLFSTVFPP | NVLFSTVFPL | 928 | 17 |
| HLA-A*02:02 | NFNVLFSTVFPP | NFNVLFSTVFPL | 2542 | 47 |
| HLA-A*02:02 | VLFSTVFPP | VLFSTVFPL | 269 | 5 |
| HLA-A*02:01 | NFNVLFSTVFPP | NFNVLFSTVFPL | 2286 | 44 |
| HLA-A*02:06 | FPPTSFGPLV | FPLTSFGPLV | 3379 | 73 |
| HLA-A*02:01 | VLFSTVFPP | VLFSTVFPL | 179 | 4 |
| HLA-A*02:06 | FNVLFSTVFPP | FNVLFSTVFPL | 1609 | 37 |
| HLA-A*02:02 | FNVLFSTVFPPT | FNVLFSTVFPLT | 4116 | 110 |
| HLA-A*02:11 | FSTVFPPTSFGPLV | FSTVFPLTSFGPLV | 6695 | 180 |
| HLA-A*02:01 | FNVLFSTVFPPTS | FNVLFSTVFPLTS | 11451 | 311 |
| HLA-A*02:06 | VLFSTVFPP | VLFSTVFPL | 368 | 10 |
| HLA-A*02:11 | ANFNVLFSTVFPPT | ANFNVLFSTVFPLT | 14639 | 409 |
| HLA-A*02:01 | FSTVFPPTSFGPLV | FSTVFPLTSFGPLV | 12596 | 425 |
| HLA-A*68:01 | PTSFGPLVR | LTSFGPLVR | 908 | 36 |
| HLA-A*02:06 | NVLFSTVFPP | NVLFSTVFPL | 1611 | 66 |
| HLA-B*15:01 | VLFSTVFPP | VLFSTVFPL | 5731 | 243 |
| HLA-B*35:01 | FPPTSFGPL | FPLTSFGPL | 304 | 13 |
| HLA-B*53:01 | FPPTSFGPL | FPLTSFGPL | 3581 | 154 |
| HLA-C*01:02 | FPPTSFGPL | FPLTSFGPL | 217 | 4855 |
| HLA-A*02:01 | CANFNVLFSTVFPP | CANFNVLFSTVFPL | 8519 | 384 |
| HLA-A*02:02 | FNVLFSTVFPPTS | FNVLFSTVFPLTS | 10920 | 497 |
| HLA-A*02:02 | FSTVFPPTSFGPLV | FSTVFPLTSFGPLV | 6958 | 322 |
| HLA-A*02:11 | VFPPTSFGPLV | VFPLTSFGPLV | 4242 | 199 |
| HLA-A*02:02 | CANFNVLFSTVFPP | CANFNVLFSTVFPL | 8183 | 391 |
| HLA-A*02:11 | NFNVLFSTVFPPT | NFNVLFSTVFPLT | 9111 | 472 |
| HLA-A*02:06 | ANFNVLFSTVFPP | ANFNVLFSTVFPL | 5191 | 304 |
| HLA-A*02:06 | NFNVLFSTVFPP | NFNVLFSTVFPL | 4220 | 274 |
| HLA-A*02:06 | FNVLFSTVFPPT | FNVLFSTVFPLT | 4868 | 388 |
| HLA-B*35:03 | FPPTSFGPL | FPLTSFGPL | 286 | 24 |
| HLA-A*03:01 | PTSFGPLVRK | LTSFGPLVRK | 554 | 47 |
| HLA-A*31:01 | PTSFGPLVR | LTSFGPLVR | 4402 | 390 |
| HLA-A*11:01 | PTSFGPLVR | LTSFGPLVR | 4068 | 379 |
| HLA-A*03:02 | PTSFGPLVRK | LTSFGPLVRK | 868 | 88 |
| HLA-B*07:02 | STVFPPTSFGPL | STVFPLTSFGPL | 777 | 79 |
| HLA-A*68:01 | PTSFGPLVRK | LTSFGPLVRK | 308 | 34 |
| HLA-B*07:02 | TVFPPTSFGPL | TVFPLTSFGPL | 1498 | 171 |
| HLA-A*02:11 | STVFPPTSFGPLV | STVFPLTSFGPLV | 2427 | 285 |
| HLA-A*02:11 | NVLFSTVFPPT | NVLFSTVFPLT | 1183 | 139 |
| HLA-A*33:03 | PTSFGPLVR | LTSFGPLVR | 3228 | 381 |
| HLA-A*26:01 | STVFPPTSF | STVFPLTSF | 786 | 95 |
| HLA-B*07:02 | VFPPTSFGPL | VFPLTSFGPL | 339 | 43 |
| HLA-A*02:01 | NVLFSTVFPPT | NVLFSTVFPLT | 2111 | 290 |
| HLA-B*42:01 | FSTVFPPTSFGPL | FSTVFPLTSFGPL | 1320 | 185 |
| HLA-B*42:01 | STVFPPTSFGPL | STVFPLTSFGPL | 738 | 111 |
| HLA-A*02:01 | VLFSTVFPPTSF | VLFSTVFPLTSF | 2225 | 342 |
| HLA-B*08:01 | FPPTSFGPL | FPLTSFGPL | 973 | 153 |
| HLA-B*42:01 | TVFPPTSFGPL | TVFPLTSFGPL | 843 | 142 |
| HLA-A*11:01 | PTSFGPLVRK | LTSFGPLVRK | 123 | 21 |
| HLA-B*42:01 | VFPPTSFGPL | VFPLTSFGPL | 274 | 47 |
| HLA-A*02:01 | VLFSTVFPPTS | VLFSTVFPLTS | 708 | 124 |
| HLA-B*07:02 | FSTVFPPTSFGPL | FSTVFPLTSFGPL | 2537 | 466 |
| HLA-B*07:02 | FPPTSFGPL | FPLTSFGPL | 42 | 8 |
| HLA-A*02:11 | VLFSTVFPPTSF | VLFSTVFPLTSF | 1019 | 195 |
| HLA-A*25:01 | STVFPPTSF | STVFPLTSF | 1533 | 296 |
| HLA-B*07:02 | FPPTSFGPLV | FPLTSFGPLV | 1131 | 236 |
| HLA-B*42:01 | FPPTSFGPL | FPLTSFGPL | 21 | 5 |
| HLA-A*26:01 | FSTVFPPTSF | FSTVFPLTSF | 1667 | 398 |
| HLA-B*51:01 | FPPTSFGPL | FPLTSFGPL | 1809 | 433 |
| HLA-A*02:11 | VLFSTVFPPTS | VLFSTVFPLTS | 309 | 74 |
| HLA-B*42:01 | FPPTSFGPLV | FPLTSFGPLV | 368 | 89 |
| HLA-A*02:02 | VLFSTVFPPTS | VLFSTVFPLTS | 1210 | 377 |
| HLA-C*03:02 | FPPTSFGPL | FPLTSFGPL | 960 | 313 |
| HLA-A*32:01 | STVFPPTSF | STVFPLTSF | 230 | 86 |
| HLA-B*58:01 | FSTVFPPTSF | FSTVFPLTSF | 88 | 33 |
| HLA-B*15:01 | STVFPPTSF | STVFPLTSF | 130 | 49 |
| HLA-A*02:01 | VLFSTVFPPT | VLFSTVFPLT | 61 | 23 |
| HLA-C*03:03 | FPPTSFGPL | FPLTSFGPL | 375 | 145 |
| HLA-C*03:04 | FPPTSFGPL | FPLTSFGPL | 375 | 145 |
| HLA-A*02:02 | VLFSTVFPPT | VLFSTVFPLT | 113 | 44 |
| HLA-C*16:01 | FSTVFPPTS | FSTVFPLTS | 1127 | 460 |
| HLA-C*14:02 | LFSTVFPPTSF | LFSTVFPLTSF | 396 | 950 |
| HLA-C*01:02 | VFPPTSFGPL | VFPLTSFGPL | 429 | 997 |
| HLA-A*02:02 | TVFPPTSFGPLV | TVFPLTSFGPLV | 789 | 343 |
| HLA-A*68:02 | STVFPPTSFGPLV | STVFPLTSFGPLV | 169 | 74 |
| HLA-A*02:11 | VLFSTVFPPT | VLFSTVFPLT | 25 | 11 |
| HLA-A*03:01 | TVFPPTSFGPLVRK | TVFPLTSFGPLVRK | 92 | 198 |
| HLA-C*14:02 | VFPPTSFGPL | VFPLTSFGPL | 47 | 99 |
| HLA-A*23:01 | LFSTVFPPTSF | LFSTVFPLTSF | 370 | 179 |
G671S
Reference CSLSHRFYRLANECAQVLSEMVMCGGSLYVKPGGTSSGDATTAYANSVFNI
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated CSLSHRFYRLANECAQVLSEMVMCGSSLYVKPGGTSSGDATTAYANSVFNI
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-B*40:02 | SEMVMCGGS | SEMVMCGSS | 1359 | 367 |
| HLA-B*50:01 | SEMVMCGGS | SEMVMCGSS | 958 | 300 |
| HLA-A*02:02 | SEMVMCGGSL | SEMVMCGSSL | 456 | 147 |
| HLA-B*40:06 | SEMVMCGGS | SEMVMCGSS | 1276 | 416 |
| HLA-B*45:01 | SEMVMCGGS | SEMVMCGSS | 173 | 59 |
| HLA-C*03:03 | MVMCGGSL | MVMCGSSL | 936 | 321 |
| HLA-C*03:04 | MVMCGGSL | MVMCGSSL | 936 | 321 |
| HLA-B*58:01 | MVMCGGSLY | MVMCGSSLY | 724 | 290 |
| HLA-B*15:03 | SEMVMCGGSL | SEMVMCGSSL | 279 | 116 |
| HLA-C*03:02 | MVMCGGSL | MVMCGSSL | 868 | 362 |
| HLA-B*15:01 | SEMVMCGGSL | SEMVMCGSSL | 437 | 183 |
| HLA-A*03:02 | MVMCGGSLY | MVMCGSSLY | 284 | 119 |
| HLA-B*15:03 | EMVMCGGSL | EMVMCGSSL | 390 | 173 |
| HLA-A*68:01 | EMVMCGGSLY | EMVMCGSSLY | 270 | 123 |
| HLA-B*35:01 | MVMCGGSLY | MVMCGSSLY | 13 | 6 |
| HLA-A*11:01 | MVMCGGSLY | MVMCGSSLY | 131 | 62 |
| HLA-B*44:02 | SEMVMCGGSL | SEMVMCGSSL | 40 | 19 |
| HLA-A*11:01 | SEMVMCGGSLY | SEMVMCGSSLY | 868 | 418 |
| HLA-B*40:02 | SEMVMCGGSL | SEMVMCGSSL | 29 | 14 |
| HLA-B*53:01 | MVMCGGSLY | MVMCGSSLY | 550 | 271 |
| HLA-A*02:02 | VLSEMVMCGG | VLSEMVMCGS | 749 | 373 |
| HLA-A*68:01 | MVMCGGSLY | MVMCGSSLY | 38 | 19 |
NSP13 protein
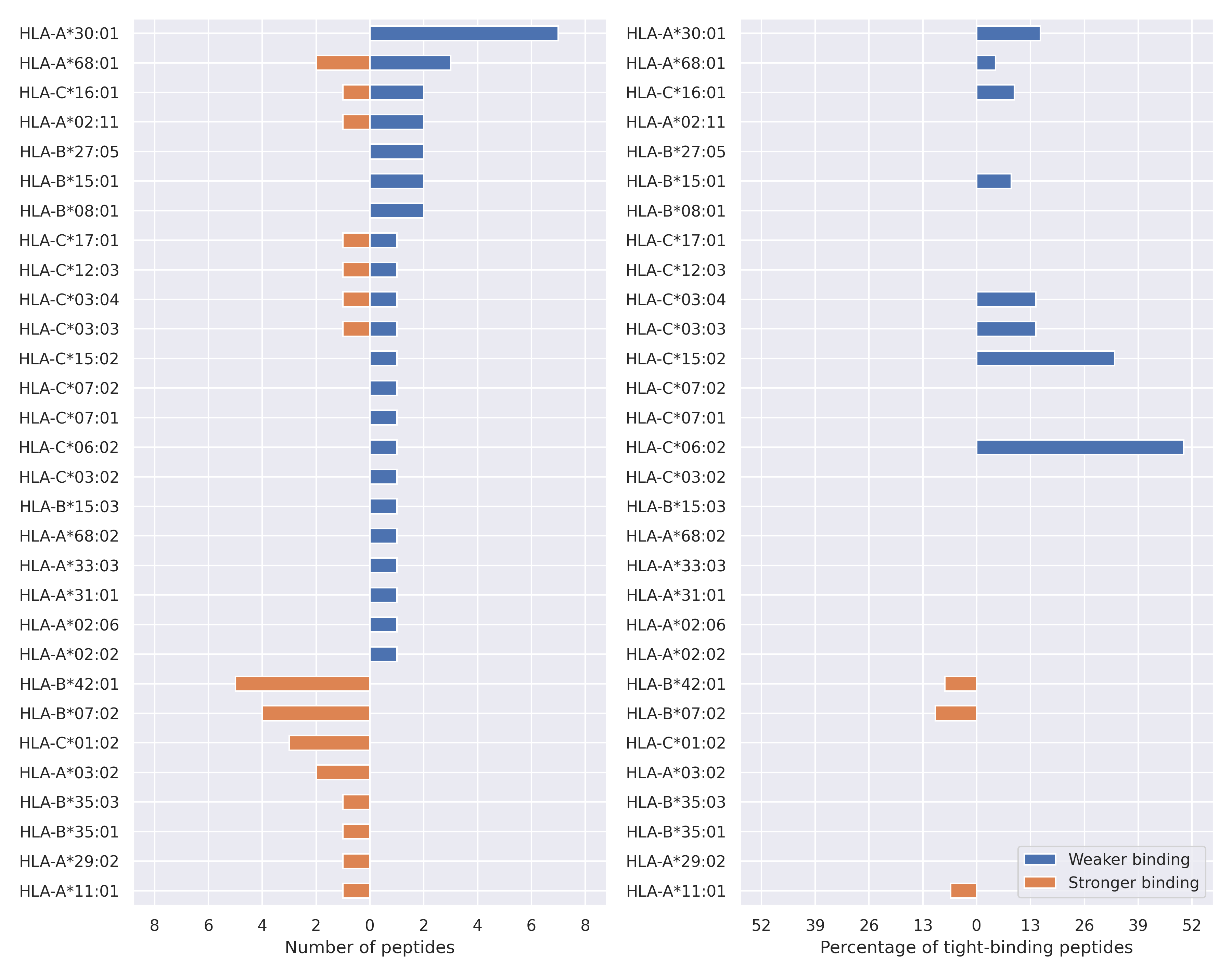
S36P
Reference QTSLRCGACIRRPFLCCKCCYDHVISTSHKLVLSVNPYVCNAPGCDVTDVT
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated QTSLRCGACIRRPFLCCKCCYDHVIPTSHKLVLSVNPYVCNAPGCDVTDVT
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-B*07:02 | ISTSHKLVL | IPTSHKLVL | 4354 | 14 |
| HLA-B*42:01 | ISTSHKLVL | IPTSHKLVL | 2286 | 9 |
| HLA-C*15:02 | ISTSHKLVL | IPTSHKLVL | 50 | 10926 |
| HLA-B*42:01 | YDHVISTSHKLVL | YDHVIPTSHKLVL | 15807 | 137 |
| HLA-B*07:02 | VISTSHKLVL | VIPTSHKLVL | 6013 | 57 |
| HLA-B*42:01 | VISTSHKLVL | VIPTSHKLVL | 4446 | 52 |
| HLA-A*68:02 | STSHKLVLSV | PTSHKLVLSV | 53 | 4422 |
| HLA-B*35:01 | ISTSHKLVL | IPTSHKLVL | 14993 | 199 |
| HLA-B*42:01 | ISTSHKLVLSV | IPTSHKLVLSV | 24838 | 330 |
| HLA-B*07:02 | YDHVISTSHKLVL | YDHVIPTSHKLVL | 21689 | 353 |
| HLA-C*16:01 | STSHKLVL | PTSHKLVL | 260 | 15946 |
| HLA-B*35:03 | ISTSHKLVL | IPTSHKLVL | 15591 | 258 |
| HLA-B*07:02 | HVISTSHKLVL | HVIPTSHKLVL | 8420 | 146 |
| HLA-C*16:01 | ISTSHKLVL | IPTSHKLVL | 9 | 518 |
| HLA-C*03:03 | ISTSHKLVL | IPTSHKLVL | 33 | 1682 |
| HLA-C*03:04 | ISTSHKLVL | IPTSHKLVL | 33 | 1682 |
| HLA-C*01:02 | VISTSHKLVL | VIPTSHKLVL | 8879 | 178 |
| HLA-B*42:01 | HVISTSHKLVL | HVIPTSHKLVL | 5488 | 142 |
| HLA-C*12:03 | ISTSHKLVL | IPTSHKLVL | 106 | 3817 |
| HLA-C*01:02 | VISTSHKL | VIPTSHKL | 16366 | 459 |
| HLA-C*01:02 | VISTSHKLV | VIPTSHKLV | 12675 | 365 |
| HLA-C*17:01 | ISTSHKLVL | IPTSHKLVL | 203 | 6625 |
| HLA-A*02:06 | STSHKLVLSV | PTSHKLVLSV | 340 | 10972 |
| HLA-C*03:02 | ISTSHKLVL | IPTSHKLVL | 83 | 2374 |
| HLA-A*02:11 | STSHKLVLSV | PTSHKLVLSV | 421 | 11088 |
| HLA-A*02:02 | STSHKLVLSV | PTSHKLVLSV | 488 | 7586 |
| HLA-A*02:11 | STSHKLVLSVNPYV | PTSHKLVLSVNPYV | 440 | 3244 |
| HLA-C*12:03 | HVISTSHKL | HVIPTSHKL | 560 | 239 |
| HLA-C*03:03 | HVISTSHKL | HVIPTSHKL | 135 | 60 |
| HLA-C*03:04 | HVISTSHKL | HVIPTSHKL | 135 | 60 |
| HLA-C*16:01 | HVISTSHKL | HVIPTSHKL | 675 | 321 |
| HLA-C*17:01 | HVISTSHKL | HVIPTSHKL | 434 | 210 |
R392C
Reference TADIVVFDEISMATNYDLSVVNARLRAKHYVYIGDPAQLPAPRTLLTKGTL
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated TADIVVFDEISMATNYDLSVVNARLCAKHYVYIGDPAQLPAPRTLLTKGTL
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-A*68:01 | DLSVVNARLR | DLSVVNARLC | 44 | 30873 |
| HLA-C*06:02 | LRAKHYVYI | LCAKHYVYI | 36 | 16159 |
| HLA-A*68:01 | LSVVNARLR | LSVVNARLC | 140 | 32767 |
| HLA-A*33:03 | DLSVVNARLR | DLSVVNARLC | 154 | 32767 |
| HLA-A*31:01 | LSVVNARLR | LSVVNARLC | 261 | 30993 |
| HLA-C*07:01 | LRAKHYVYI | LCAKHYVYI | 133 | 15165 |
| HLA-B*27:05 | LRAKHYVYI | LCAKHYVYI | 379 | 32767 |
| HLA-A*30:01 | RLRAKHYVY | RLCAKHYVY | 27 | 1483 |
| HLA-B*27:05 | RLRAKHYVYI | RLCAKHYVYI | 208 | 10096 |
| HLA-C*07:02 | LRAKHYVYI | LCAKHYVYI | 433 | 17614 |
| HLA-A*30:01 | RAKHYVYIG | CAKHYVYIG | 76 | 2630 |
| HLA-A*68:01 | TNYDLSVVNARLR | TNYDLSVVNARLC | 477 | 15989 |
| HLA-A*30:01 | RLRAKHYV | RLCAKHYV | 218 | 5721 |
| HLA-A*30:01 | RLRAKHYVYI | RLCAKHYVYI | 17 | 427 |
| HLA-A*30:01 | RAKHYVYIGDPA | CAKHYVYIGDPA | 188 | 4601 |
| HLA-A*30:01 | RAKHYVYI | CAKHYVYI | 211 | 4134 |
| HLA-B*08:01 | RLRAKHYV | RLCAKHYV | 363 | 6671 |
| HLA-A*30:01 | ARLRAKHYVYI | ARLCAKHYVYI | 422 | 6081 |
| HLA-B*08:01 | RLRAKHYVYI | RLCAKHYVYI | 230 | 2547 |
| HLA-B*15:03 | LRAKHYVY | LCAKHYVY | 395 | 4119 |
| HLA-A*29:02 | RLRAKHYVY | RLCAKHYVY | 535 | 101 |
| HLA-A*02:11 | RLRAKHYV | RLCAKHYV | 1483 | 352 |
| HLA-A*68:01 | VVNARLRAK | VVNARLCAK | 1428 | 381 |
| HLA-A*11:01 | VVNARLRAK | VVNARLCAK | 145 | 44 |
| HLA-A*03:02 | VVNARLRAK | VVNARLCAK | 257 | 79 |
| HLA-B*15:01 | ARLRAKHYVY | ARLCAKHYVY | 402 | 1223 |
| HLA-B*15:01 | RLRAKHYVY | RLCAKHYVY | 49 | 133 |
| HLA-A*68:01 | SVVNARLRAK | SVVNARLCAK | 158 | 63 |
| HLA-A*03:02 | SVVNARLRAK | SVVNARLCAK | 196 | 86 |
NSP14 protein
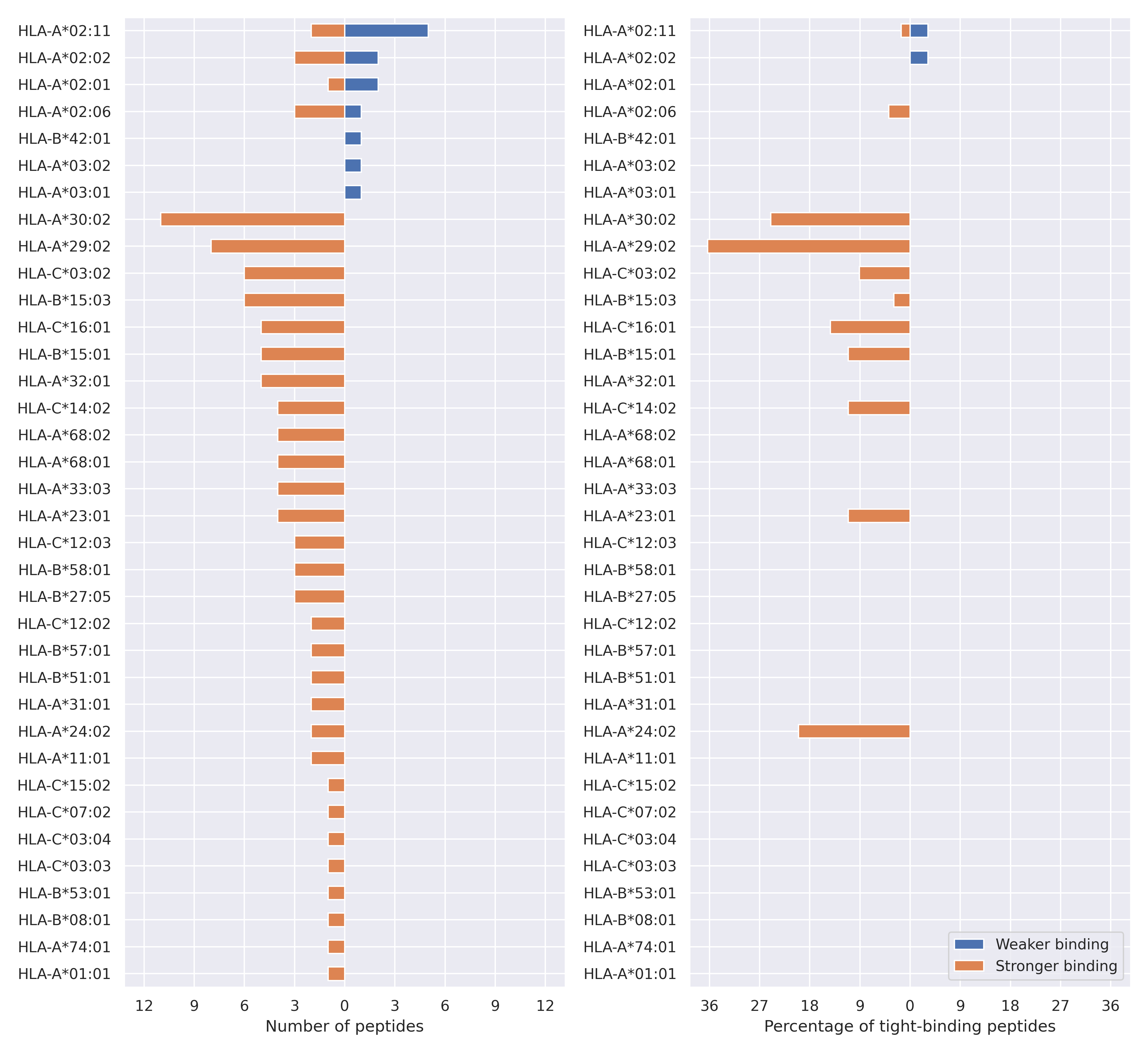
I42V
Reference GLHPTQAPTHLSVDTKFKTEGLCVDIPGIPKDMTYRRLISMMGFKMNYQVN
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated GLHPTQAPTHLSVDTKFKTEGLCVDVPGIPKDMTYRRLISMMGFKMNYQVN
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-A*68:01 | DIPGIPKDMTYRR | DVPGIPKDMTYRR | 570 | 271 |
L157F
Reference DFSRVSAKPPPGDQFKHLIPLMYKGLPWNVVRIKIVQMLSDTLKNLSDRVV
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated DFSRVSAKPPPGDQFKHLIPLMYKGFPWNVVRIKIVQMLSDTLKNLSDRVV
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-A*02:11 | GLPWNVVRI | GFPWNVVRI | 19 | 5788 |
| HLA-A*02:02 | GLPWNVVRI | GFPWNVVRI | 96 | 10946 |
| HLA-A*02:11 | HLIPLMYKGL | HLIPLMYKGF | 26 | 2716 |
| HLA-A*02:02 | HLIPLMYKGL | HLIPLMYKGF | 31 | 1879 |
| HLA-A*02:01 | GLPWNVVRI | GFPWNVVRI | 390 | 18925 |
| HLA-A*02:11 | YKGLPWNVVRI | YKGFPWNVVRI | 224 | 10133 |
| HLA-A*02:01 | HLIPLMYKGL | HLIPLMYKGF | 114 | 4786 |
| HLA-A*02:11 | MYKGLPWNVVRI | MYKGFPWNVVRI | 491 | 11790 |
| HLA-A*03:01 | GLPWNVVRIK | GFPWNVVRIK | 153 | 3511 |
| HLA-A*02:11 | KGLPWNVVRI | KGFPWNVVRI | 167 | 3506 |
| HLA-B*15:01 | HLIPLMYKGL | HLIPLMYKGF | 5031 | 306 |
| HLA-A*02:06 | HLIPLMYKGL | HLIPLMYKGF | 153 | 2257 |
| HLA-A*68:01 | YKGLPWNVVR | YKGFPWNVVR | 2055 | 178 |
| HLA-A*03:02 | GLPWNVVRIK | GFPWNVVRIK | 361 | 3138 |
| HLA-B*42:01 | IPLMYKGL | IPLMYKGF | 280 | 2062 |
| HLA-A*68:01 | MYKGLPWNVVR | MYKGFPWNVVR | 1761 | 328 |
| HLA-A*74:01 | KGLPWNVVR | KGFPWNVVR | 1335 | 361 |
| HLA-B*53:01 | LPWNVVRIKI | FPWNVVRIKI | 585 | 163 |
| HLA-A*33:03 | YKGLPWNVVR | YKGFPWNVVR | 972 | 276 |
| HLA-B*08:01 | LPWNVVRIKIVQM | FPWNVVRIKIVQM | 1695 | 496 |
| HLA-A*33:03 | KGLPWNVVR | KGFPWNVVR | 1505 | 496 |
| HLA-A*31:01 | YKGLPWNVVR | YKGFPWNVVR | 453 | 159 |
| HLA-A*32:01 | PLMYKGLPW | PLMYKGFPW | 1107 | 451 |
| HLA-C*07:02 | MYKGLPWNV | MYKGFPWNV | 1075 | 447 |
| HLA-A*31:01 | KGLPWNVVR | KGFPWNVVR | 24 | 10 |
| HLA-A*32:01 | LMYKGLPW | LMYKGFPW | 452 | 193 |
| HLA-A*33:03 | PLMYKGLPWNVVR | PLMYKGFPWNVVR | 800 | 358 |
| HLA-A*02:02 | LIPLMYKGLPWNV | LIPLMYKGFPWNV | 904 | 412 |
| HLA-C*14:02 | MYKGLPWNV | MYKGFPWNV | 324 | 148 |
| HLA-A*33:03 | MYKGLPWNVVRI | MYKGFPWNVVRI | 672 | 318 |
| HLA-B*51:01 | LPWNVVRIKI | FPWNVVRIKI | 510 | 247 |
| HLA-B*51:01 | LPWNVVRI | FPWNVVRI | 221 | 109 |
| HLA-A*02:11 | LIPLMYKGLPWNV | LIPLMYKGFPWNV | 255 | 127 |
| HLA-A*32:01 | IPLMYKGLPW | IPLMYKGFPW | 230 | 115 |
D222Y
Reference YFVKIGPERTCCLCDRRATCFSTASDTYACWHHSIGFDYVYNPFMIDVQQW
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated YFVKIGPERTCCLCDRRATCFSTASYTYACWHHSIGFDYVYNPFMIDVQQW
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-B*15:03 | RATCFSTASD | RATCFSTASY | 19966 | 19 |
| HLA-A*29:02 | ATCFSTASD | ATCFSTASY | 32767 | 45 |
| HLA-B*15:01 | RATCFSTASD | RATCFSTASY | 21694 | 35 |
| HLA-A*30:02 | RATCFSTASD | RATCFSTASY | 10071 | 21 |
| HLA-A*30:02 | ATCFSTASD | ATCFSTASY | 10771 | 24 |
| HLA-C*03:02 | RATCFSTASD | RATCFSTASY | 28224 | 65 |
| HLA-C*16:01 | RATCFSTASD | RATCFSTASY | 25570 | 63 |
| HLA-A*11:01 | ATCFSTASD | ATCFSTASY | 25150 | 71 |
| HLA-B*15:03 | RRATCFSTASD | RRATCFSTASY | 23667 | 71 |
| HLA-B*15:01 | ATCFSTASD | ATCFSTASY | 23432 | 75 |
| HLA-A*30:02 | RRATCFSTASD | RRATCFSTASY | 18128 | 75 |
| HLA-A*02:11 | DTYACWHHSI | YTYACWHHSI | 4030 | 18 |
| HLA-B*15:03 | ATCFSTASD | ATCFSTASY | 27683 | 132 |
| HLA-A*01:01 | ATCFSTASD | ATCFSTASY | 28819 | 179 |
| HLA-A*02:06 | DTYACWHHSI | YTYACWHHSI | 6190 | 39 |
| HLA-C*16:01 | ATCFSTASD | ATCFSTASY | 29018 | 194 |
| HLA-C*03:02 | ATCFSTASD | ATCFSTASY | 32551 | 243 |
| HLA-A*29:02 | RATCFSTASD | RATCFSTASY | 32767 | 309 |
| HLA-A*11:01 | RATCFSTASD | RATCFSTASY | 28360 | 296 |
| HLA-C*15:02 | DTYACWHHSI | YTYACWHHSI | 11630 | 139 |
| HLA-A*30:02 | DRRATCFSTASD | DRRATCFSTASY | 28594 | 353 |
| HLA-C*12:03 | DTYACWHHSI | YTYACWHHSI | 7516 | 97 |
| HLA-C*12:02 | RATCFSTASD | RATCFSTASY | 27331 | 383 |
| HLA-A*32:01 | DTYACWHHSI | YTYACWHHSI | 5967 | 84 |
| HLA-A*02:02 | DTYACWHHSI | YTYACWHHSI | 6590 | 93 |
| HLA-C*03:02 | DTYACWHHSI | YTYACWHHSI | 15619 | 227 |
| HLA-B*15:01 | RRATCFSTASD | RRATCFSTASY | 32231 | 480 |
| HLA-A*02:01 | DTYACWHHSI | YTYACWHHSI | 13425 | 203 |
| HLA-A*68:01 | ATCFSTASD | ATCFSTASY | 23506 | 434 |
| HLA-A*02:06 | DTYACWHHS | YTYACWHHS | 13200 | 247 |
| HLA-C*16:01 | DTYACWHHSI | YTYACWHHSI | 8864 | 174 |
| HLA-C*03:03 | DTYACWHHSI | YTYACWHHSI | 20873 | 489 |
| HLA-C*03:04 | DTYACWHHSI | YTYACWHHSI | 20873 | 489 |
| HLA-B*27:05 | RRATCFSTASD | RRATCFSTASY | 3385 | 83 |
| HLA-A*29:02 | DTYACWHHSIGFDY | YTYACWHHSIGFDY | 1814 | 45 |
| HLA-C*14:02 | DTYACWHHSI | YTYACWHHSI | 3032 | 81 |
| HLA-A*23:01 | SDTYACWHHSIGF | SYTYACWHHSIGF | 4778 | 146 |
| HLA-A*24:02 | SDTYACWHHSIGF | SYTYACWHHSIGF | 6582 | 227 |
| HLA-B*15:01 | DTYACWHHSIGF | YTYACWHHSIGF | 11353 | 422 |
| HLA-C*03:02 | DTYACWHHSIGF | YTYACWHHSIGF | 9267 | 390 |
| HLA-B*15:03 | DTYACWHHSIGF | YTYACWHHSIGF | 7332 | 309 |
| HLA-A*30:02 | ATCFSTASDT | ATCFSTASYT | 9799 | 466 |
| HLA-A*24:02 | DTYACWHHSI | YTYACWHHSI | 625 | 30 |
| HLA-A*23:01 | DTYACWHHSI | YTYACWHHSI | 782 | 43 |
| HLA-B*58:01 | RATCFSTASD | RATCFSTASY | 2644 | 154 |
| HLA-A*29:02 | DTYACWHHSIGF | YTYACWHHSIGF | 6004 | 449 |
| HLA-B*57:01 | ASDTYACW | ASYTYACW | 2121 | 196 |
| HLA-A*29:02 | TCFSTASDTY | TCFSTASYTY | 364 | 36 |
| HLA-A*32:01 | STASDTYACW | STASYTYACW | 952 | 95 |
| HLA-C*12:03 | FSTASDTYA | FSTASYTYA | 1391 | 144 |
| HLA-A*30:02 | DTYACWHHSIGFDY | YTYACWHHSIGFDY | 3359 | 349 |
| HLA-A*02:06 | FSTASDTYA | FSTASYTYA | 1223 | 128 |
| HLA-A*29:02 | CFSTASDTY | CFSTASYTY | 102 | 12 |
| HLA-A*30:02 | ASDTYACWH | ASYTYACWH | 3792 | 472 |
| HLA-C*16:01 | FSTASDTY | FSTASYTY | 306 | 44 |
| HLA-A*68:02 | FSTASDTYA | FSTASYTYA | 409 | 59 |
| HLA-A*29:02 | RATCFSTASDTY | RATCFSTASYTY | 2065 | 342 |
| HLA-B*58:01 | ASDTYACW | ASYTYACW | 723 | 121 |
| HLA-C*16:01 | FSTASDTYA | FSTASYTYA | 1130 | 206 |
| HLA-C*12:03 | FSTASDTY | FSTASYTY | 2346 | 430 |
| HLA-B*58:01 | RATCFSTASDTY | RATCFSTASYTY | 816 | 155 |
| HLA-A*29:02 | ATCFSTASDTY | ATCFSTASYTY | 1102 | 217 |
| HLA-C*03:02 | FSTASDTY | FSTASYTY | 228 | 46 |
| HLA-C*12:02 | FSTASDTYA | FSTASYTYA | 2240 | 472 |
| HLA-A*02:02 | FSTASDTYA | FSTASYTYA | 1752 | 373 |
| HLA-B*15:03 | FSTASDTY | FSTASYTY | 995 | 231 |
| HLA-A*30:02 | CFSTASDTY | CFSTASYTY | 482 | 115 |
| HLA-C*14:02 | TCFSTASDTY | TCFSTASYTY | 1708 | 408 |
| HLA-C*03:02 | FSTASDTYA | FSTASYTYA | 850 | 229 |
| HLA-B*27:05 | RRATCFSTASDT | RRATCFSTASYT | 1375 | 371 |
| HLA-A*68:02 | STASDTYAC | STASYTYAC | 743 | 201 |
| HLA-A*30:02 | TCFSTASDTY | TCFSTASYTY | 455 | 146 |
| HLA-C*14:02 | CFSTASDTY | CFSTASYTY | 96 | 31 |
| HLA-A*30:02 | RRATCFSTASDTY | RRATCFSTASYTY | 605 | 215 |
| HLA-A*23:01 | DTYACWHHSIGF | YTYACWHHSIGF | 939 | 364 |
| HLA-A*68:02 | STASDTYACWHHSI | STASYTYACWHHSI | 905 | 362 |
| HLA-B*15:03 | CFSTASDTY | CFSTASYTY | 783 | 338 |
| HLA-A*23:01 | SDTYACWHHSI | SYTYACWHHSI | 274 | 119 |
| HLA-A*30:02 | RATCFSTASDTY | RATCFSTASYTY | 228 | 100 |
| HLA-A*68:02 | DTYACWHHSI | YTYACWHHSI | 31 | 14 |
| HLA-B*27:05 | RRATCFSTASDTY | RRATCFSTASYTY | 260 | 118 |
| HLA-B*57:01 | TASDTYACW | TASYTYACW | 187 | 87 |
NSP15 protein
T112I
Reference DYKRDAPAHISTIGVCSMTDIAKKPTETICAPLTVFFDGRVDGQVDLFRNA
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated DYKRDAPAHISTIGVCSMTDIAKKPIETICAPLTVFFDGRVDGQVDLFRNA
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-A*02:02 | SMTDIAKKPT | SMTDIAKKPI | 3353 | 375 |
| HLA-A*68:02 | TETICAPLTV | IETICAPLTV | 64 | 161 |
| HLA-C*03:03 | IAKKPTETI | IAKKPIETI | 497 | 1187 |
| HLA-C*03:04 | IAKKPTETI | IAKKPIETI | 497 | 1187 |
| HLA-A*02:11 | SMTDIAKKPTETI | SMTDIAKKPIETI | 560 | 263 |