Summary
This strain contains 26 protein-level mutations:
- 9 mutations in Spike protein: T95I, 144-145 YY->SN, R346K, E484K, N501Y, D614G, Q677H, P681H, D950N
- 2 mutations in N protein: T205I, Q241K
- 1 mutation in NS3 protein: Q57H
- 3 mutations in NS8 protein: T11K, P38S, S67F
- 1 mutation in NS9b protein: R32L
- 1 mutation in NS9c protein: L52F
- 3 mutations in NSP3 protein: T237A, T720I, E997K
- 1 mutation in NSP4 protein: T492I
- 1 mutation in NSP6 protein: Q160R
- 1 mutation in NSP12 protein: P323L
- 2 mutations in NSP13 protein: P419S, L581F
- 1 mutation in NSP14 protein: V125F
We identified all possible linear viral peptides affected by these mutations. Whenever it was possible, we matched the reference peptide with the mutated one. For example, D -> L mutation transformed SDNGPQNQR to SLNGPQNQR. Cases when it was not meaningful included deletions and insertions at the flanks of the peptide, e.g., HV deletion in NVTWFHAIHV peptide.
Then, we predicted binding affinities between the selected peptides and frequent HLA alleles. Predictions were made with NetMHCpan-4.1 and NetMHCIIpan-4.0. The binding affinities were classifies into three groups:
- Tight binding (IC50 affinity ≤ 50 nM)
- Moderate binding (50 nM < IC50 affinity ≤ 500 nM)
- Weak/no binding (IC50 affinity > 500 nM)
Here we report HLA-peptide interactions whose affinity was altered by at least two folds. Note that mutations with empty set of altered interactions are not showed.
The total number of interactions between HLA-DQA1*01:02/DQB1*06:09 and peptides affected by the mutations
Weaker binding was not found, while stronger binding was found for 37 entries. There is no tight binders for the HLA-DQA1*01:02/DQB1*06:09 in the reference immunopeptidome (IC50 affinity ≤ 50 nM). To avoid possible divisions by zero, we used + 1 regularization term in denominators when calculating percentages.
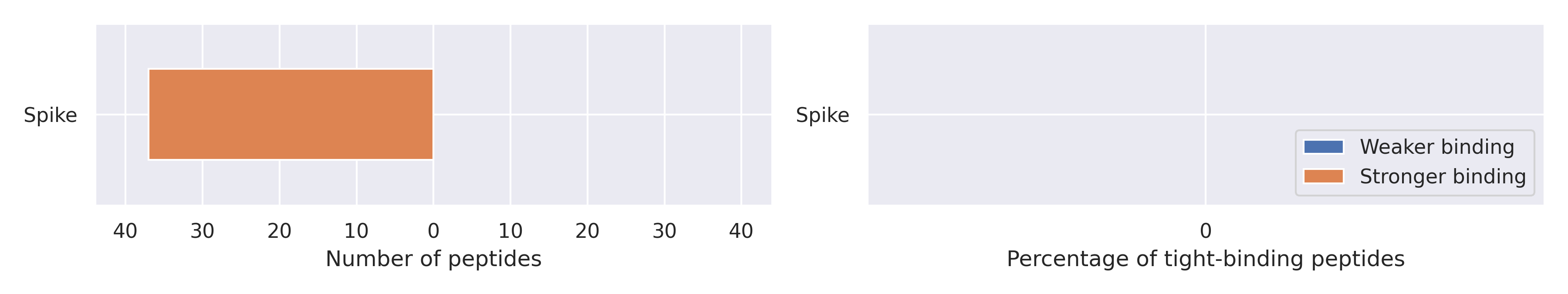
Spike protein
T95I
Reference VSGTNGTKRFDNPVLPFNDGVYFASTEKSNIIRGWIFGTTLDSKTQSLLIV
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated VSGTNGTKRFDNPVLPFNDGVYFASIEKSNIIRGWIFGTTLDSKTQSLLIV
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-DQA1*01:02/DQB1*06:09 | FNDGVYFASTEKSNI | FNDGVYFASIEKSNI | 922 | 339 |
| HLA-DQA1*01:02/DQB1*06:09 | PFNDGVYFASTEKSN | PFNDGVYFASIEKSN | 1083 | 402 |
| HLA-DQA1*01:02/DQB1*06:09 | FNDGVYFASTEKSNIIRG | FNDGVYFASIEKSNIIRG | 1320 | 493 |
| HLA-DQA1*01:02/DQB1*06:09 | PFNDGVYFASTEKSNIIRG | PFNDGVYFASIEKSNIIRG | 795 | 311 |
| HLA-DQA1*01:02/DQB1*06:09 | LPFNDGVYFASTEKS | LPFNDGVYFASIEKS | 922 | 364 |
| HLA-DQA1*01:02/DQB1*06:09 | NDGVYFASTEKSNIIRGWI | NDGVYFASIEKSNIIRGWI | 886 | 352 |
| HLA-DQA1*01:02/DQB1*06:09 | FNDGVYFASTEKSNIIRGW | FNDGVYFASIEKSNIIRGW | 809 | 325 |
| HLA-DQA1*01:02/DQB1*06:09 | NDGVYFASTEKSNIIRG | NDGVYFASIEKSNIIRG | 904 | 368 |
| HLA-DQA1*01:02/DQB1*06:09 | DGVYFASTEKSNIIRGW | DGVYFASIEKSNIIRGW | 951 | 390 |
| HLA-DQA1*01:02/DQB1*06:09 | NDGVYFASTEKSNII | NDGVYFASIEKSNII | 807 | 338 |
| HLA-DQA1*01:02/DQB1*06:09 | LPFNDGVYFASTEKSNII | LPFNDGVYFASIEKSNII | 1065 | 449 |
| HLA-DQA1*01:02/DQB1*06:09 | LPFNDGVYFASTEKSN | LPFNDGVYFASIEKSN | 786 | 334 |
| HLA-DQA1*01:02/DQB1*06:09 | LPFNDGVYFASTEKSNI | LPFNDGVYFASIEKSNI | 804 | 342 |
| HLA-DQA1*01:02/DQB1*06:09 | PFNDGVYFASTEKSNII | PFNDGVYFASIEKSNII | 754 | 321 |
| HLA-DQA1*01:02/DQB1*06:09 | FNDGVYFASTEKSNIIR | FNDGVYFASIEKSNIIR | 788 | 338 |
| HLA-DQA1*01:02/DQB1*06:09 | DGVYFASTEKSNIIRG | DGVYFASIEKSNIIRG | 786 | 339 |
| HLA-DQA1*01:02/DQB1*06:09 | PFNDGVYFASTEKSNI | PFNDGVYFASIEKSNI | 740 | 321 |
| HLA-DQA1*01:02/DQB1*06:09 | GVYFASTEKSNIIRG | GVYFASIEKSNIIRG | 1023 | 444 |
| HLA-DQA1*01:02/DQB1*06:09 | VLPFNDGVYFASTEKSN | VLPFNDGVYFASIEKSN | 845 | 370 |
| HLA-DQA1*01:02/DQB1*06:09 | FNDGVYFASTEKSNIIRGWI | FNDGVYFASIEKSNIIRGWI | 635 | 286 |
| HLA-DQA1*01:02/DQB1*06:09 | DGVYFASTEKSNIIRGWIF | DGVYFASIEKSNIIRGWIF | 799 | 361 |
| HLA-DQA1*01:02/DQB1*06:09 | FNDGVYFASTEKSNII | FNDGVYFASIEKSNII | 615 | 278 |
| HLA-DQA1*01:02/DQB1*06:09 | DGVYFASTEKSNIIR | DGVYFASIEKSNIIR | 907 | 411 |
| HLA-DQA1*01:02/DQB1*06:09 | LPFNDGVYFASTEKSNIIR | LPFNDGVYFASIEKSNIIR | 680 | 309 |
| HLA-DQA1*01:02/DQB1*06:09 | PFNDGVYFASTEKSNIIRGW | PFNDGVYFASIEKSNIIRGW | 631 | 288 |
| HLA-DQA1*01:02/DQB1*06:09 | GVYFASTEKSNIIRGWI | GVYFASIEKSNIIRGWI | 1004 | 462 |
| HLA-DQA1*01:02/DQB1*06:09 | LPFNDGVYFASTEKSNIIRG | LPFNDGVYFASIEKSNIIRG | 585 | 272 |
| HLA-DQA1*01:02/DQB1*06:09 | VLPFNDGVYFASTEKSNI | VLPFNDGVYFASIEKSNI | 1045 | 489 |
| HLA-DQA1*01:02/DQB1*06:09 | PVLPFNDGVYFASTEKS | PVLPFNDGVYFASIEKS | 824 | 386 |
| HLA-DQA1*01:02/DQB1*06:09 | VLPFNDGVYFASTEKS | VLPFNDGVYFASIEKS | 694 | 326 |
| HLA-DQA1*01:02/DQB1*06:09 | GVYFASTEKSNIIRGWIFG | GVYFASIEKSNIIRGWIFG | 960 | 454 |
| HLA-DQA1*01:02/DQB1*06:09 | NDGVYFASTEKSNIIR | NDGVYFASIEKSNIIR | 711 | 337 |
| HLA-DQA1*01:02/DQB1*06:09 | PVLPFNDGVYFASTEKSNI | PVLPFNDGVYFASIEKSNI | 679 | 324 |
| HLA-DQA1*01:02/DQB1*06:09 | GVYFASTEKSNIIRGW | GVYFASIEKSNIIRGW | 830 | 402 |
| HLA-DQA1*01:02/DQB1*06:09 | NPVLPFNDGVYFASTEKSN | NPVLPFNDGVYFASIEKSN | 738 | 358 |
| HLA-DQA1*01:02/DQB1*06:09 | VLPFNDGVYFASTEKSNII | VLPFNDGVYFASIEKSNII | 596 | 291 |
| HLA-DQA1*01:02/DQB1*06:09 | NDGVYFASTEKSNIIRGWIF | NDGVYFASIEKSNIIRGWIF | 636 | 318 |