Summary
This strain contains 18 protein-level mutations:
- 9 mutations in Spike protein: D80A, D215G, LLA 241-243 deletion, A260X, K417N, E484K, N501Y, D614G, A701V
- 1 mutation in N protein: T205I
- 1 mutation in E protein: P71L
- 2 mutations in NS3 protein: Q57H, S171L
- 1 mutation in NS9c protein: L52F
- 1 mutation in NSP2 protein: T85I
- 1 mutation in NSP3 protein: K837N
- 1 mutation in NSP6 protein: SGF 106-108 deletion
- 1 mutation in NSP12 protein: P323L
We identified all possible linear viral peptides affected by these mutations. Whenever it was possible, we matched the reference peptide with the mutated one. For example, D -> L mutation transformed SDNGPQNQR to SLNGPQNQR. Cases when it was not meaningful included deletions and insertions at the flanks of the peptide, e.g., HV deletion in NVTWFHAIHV peptide.
Then, we predicted binding affinities between the selected peptides and frequent HLA alleles. Predictions were made with NetMHCpan-4.1 and NetMHCIIpan-4.0. The binding affinities were classifies into three groups:
- Tight binding (IC50 affinity ≤ 50 nM)
- Moderate binding (50 nM < IC50 affinity ≤ 500 nM)
- Weak/no binding (IC50 affinity > 500 nM)
Here we report HLA-peptide interactions whose affinity was altered by at least two folds. Note that mutations with empty set of altered interactions are not showed.
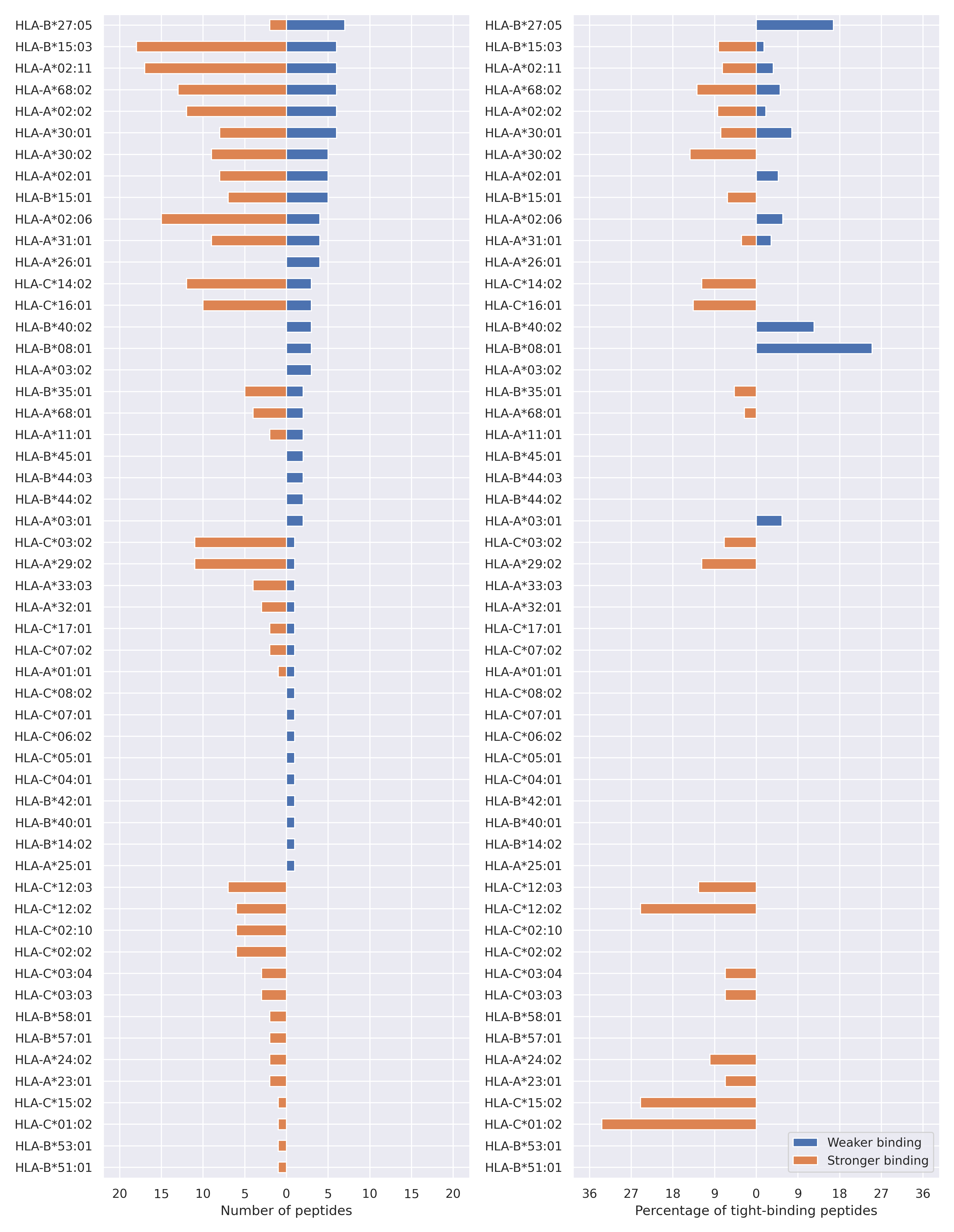
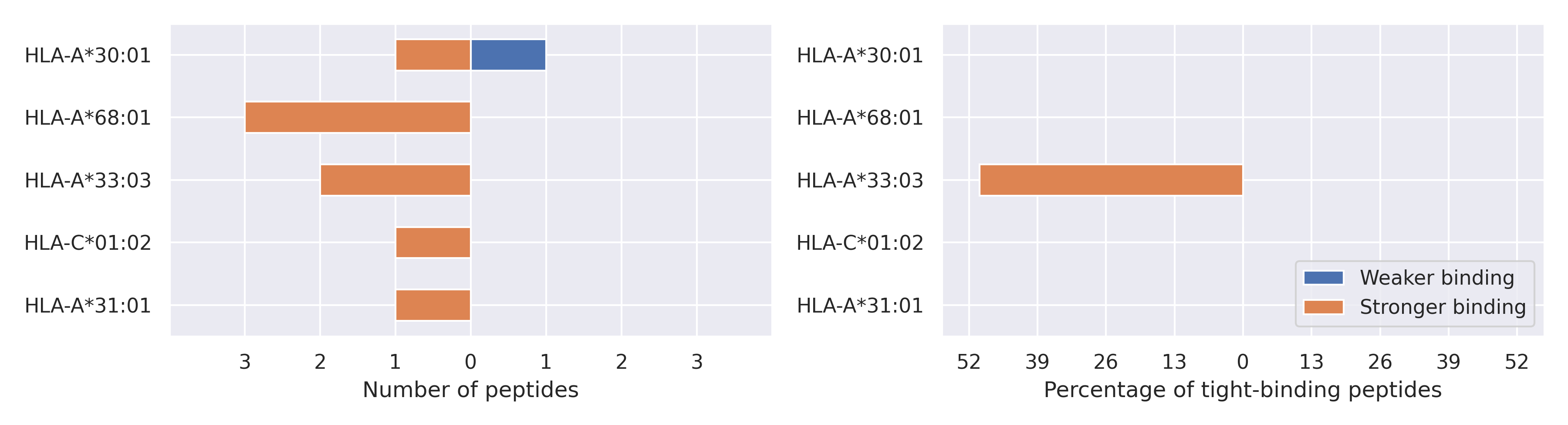
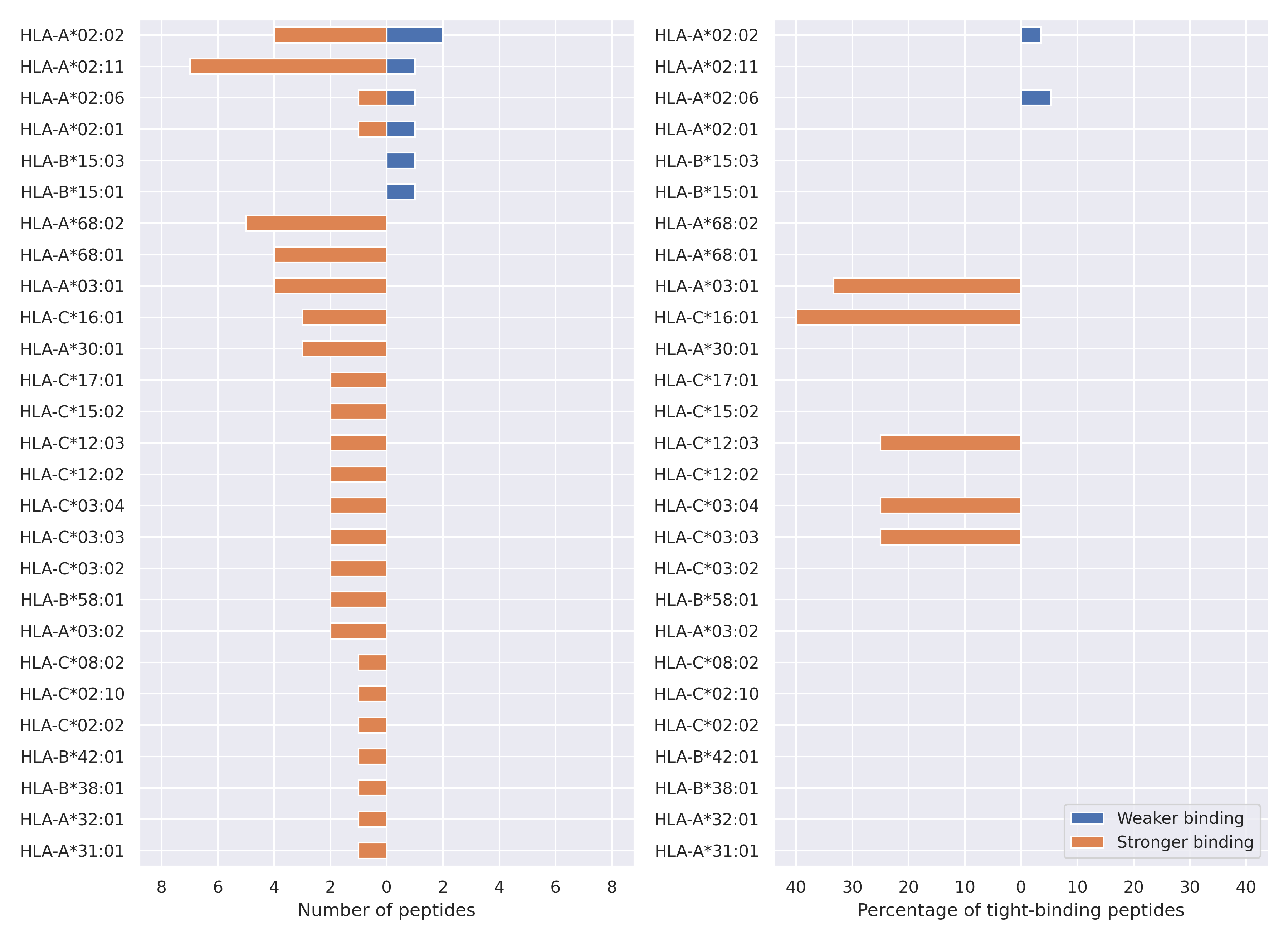
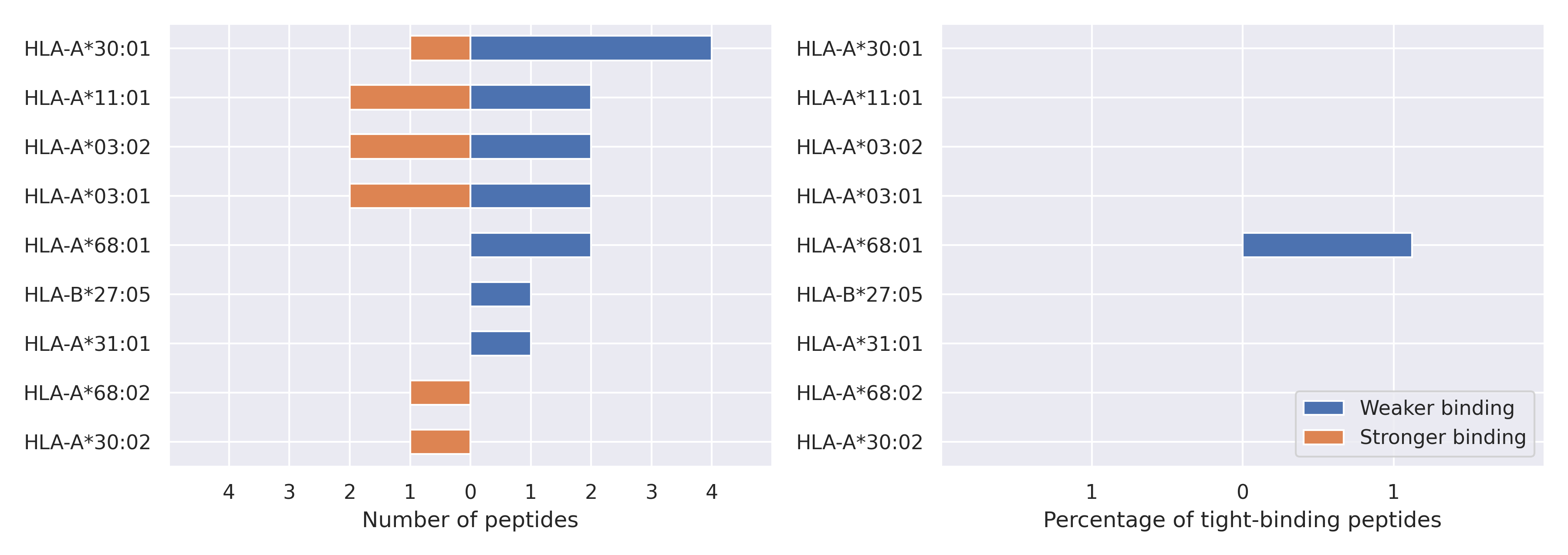
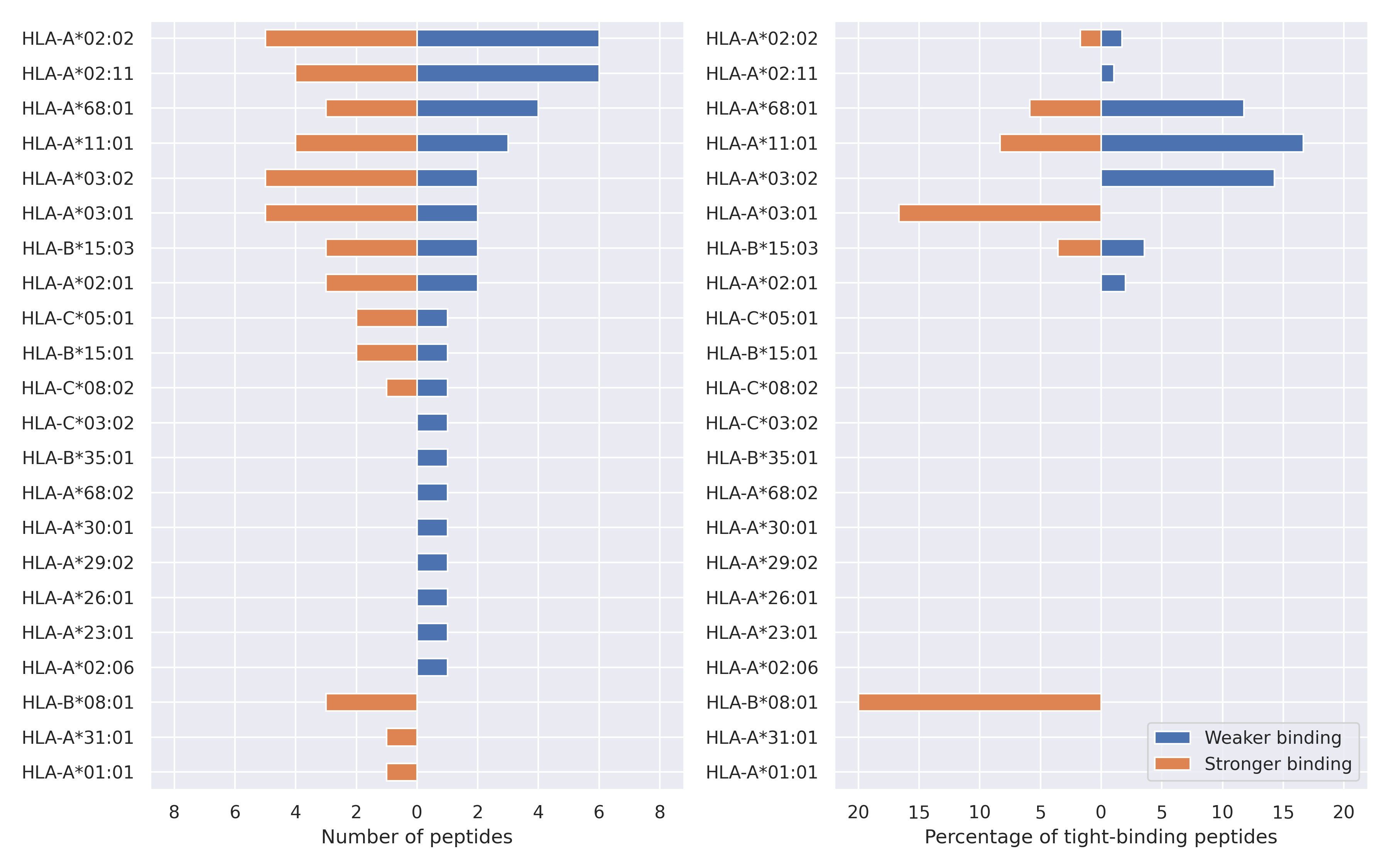
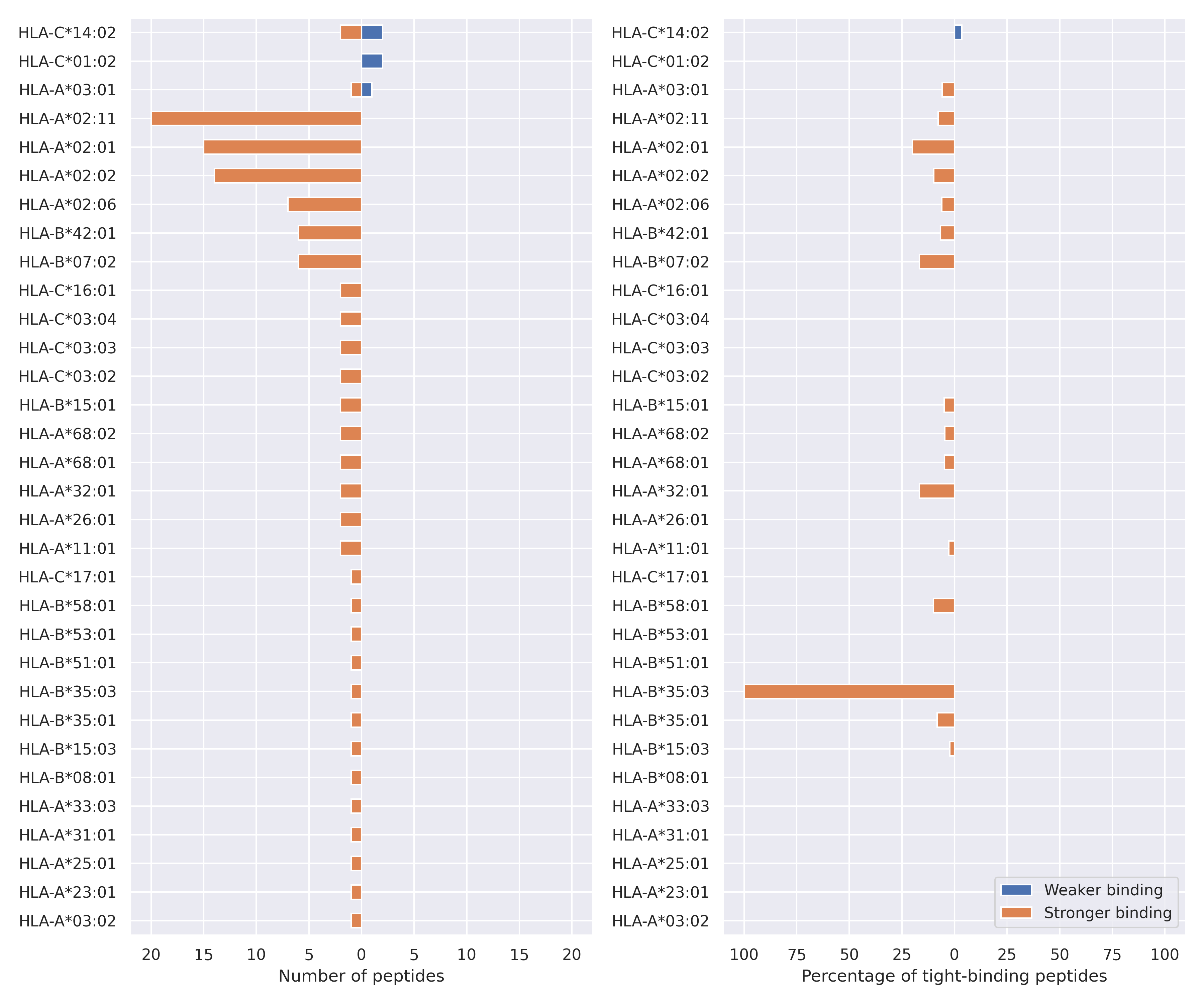
The total number of HLA-peptide interactions affected by the mutations
For each allele we calculated the number of peptides with increased and decreased affinitiy (two or more times). The absolute number of such peptides is showed on the left side of the figure (blue bars — weaker binding, orange bars — stronger).
While absolute numbers are of interest, they do not reflect the initial number of tighly binding peptides for the specific allele. Roughly speaking, if an allele had high number of tight binders, then vanished affinity of several peptides will not affect much the integral ability of peptide presentation. To the contrary, low number of vanished peptides could be important if the particular allele initially had narrow epitope repertoire. To account for this effect, we normalized the absolute numbers of new/vanished tight binders (IC50 affinity ≤ 50 nM) by the total number of tight binders for each allele. To avoid possible divisions by zero, we used + 1 regularization term in all denominators. For example, if some allele had no tight binders in the reference genome, and three new tight binders appeared as a result of the mutation, the relative increase will be 300%. The results are showed in the right part of the figure. Protein-specific plots are listed below.
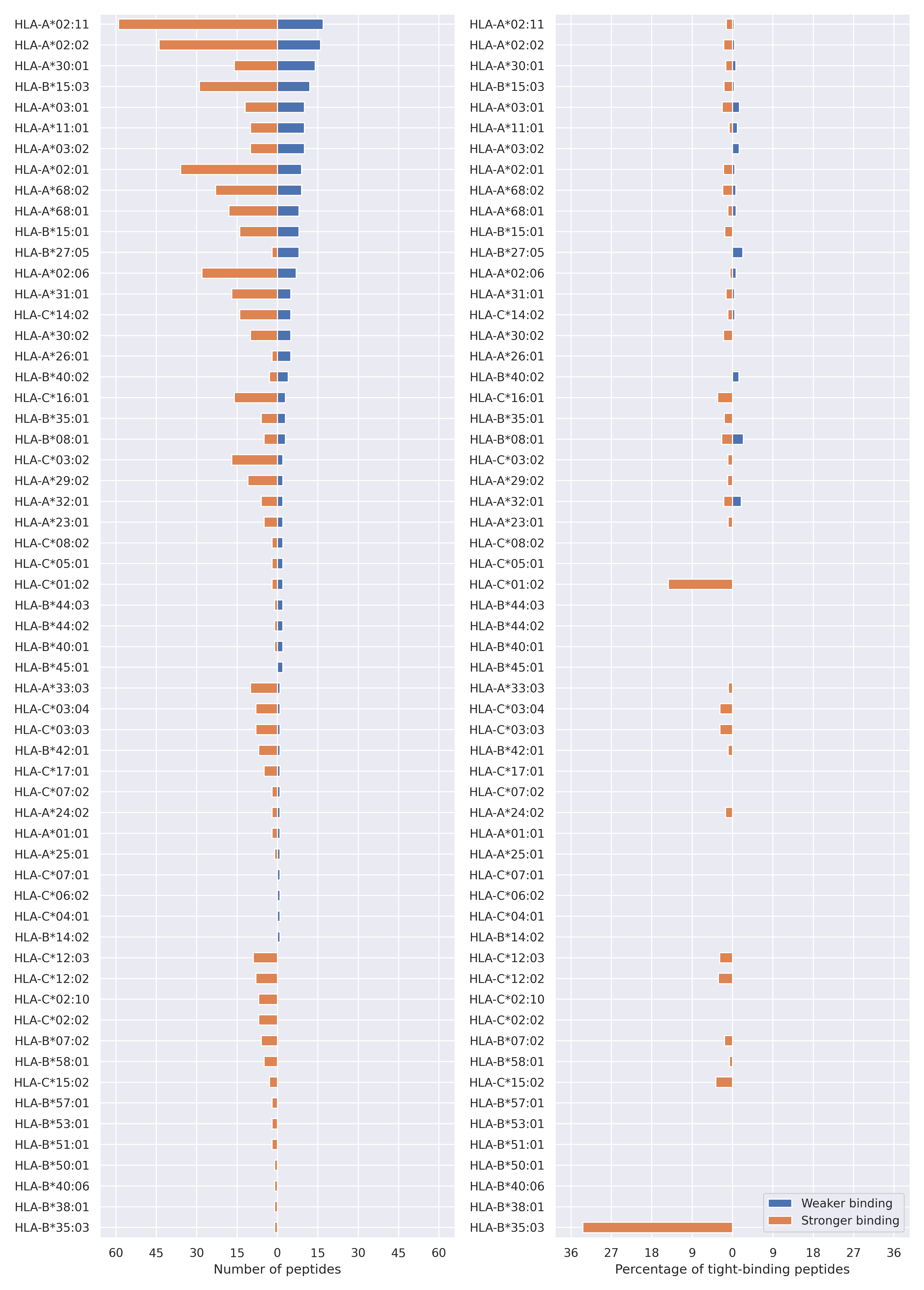
Spike protein
D80A
Reference FLPFFSNVTWFHAIHVSGTNGTKRFDNPVLPFNDGVYFASTEKSNIIRGWI
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated FLPFFSNVTWFHAIHVSGTNGTKRFANPVLPFNDGVYFASTEKSNIIRGWI
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-C*03:02 | FDNPVLPF | FANPVLPF | 4198 | 5 |
| HLA-C*16:01 | FDNPVLPF | FANPVLPF | 9054 | 16 |
| HLA-C*03:03 | FDNPVLPF | FANPVLPF | 15346 | 35 |
| HLA-C*03:04 | FDNPVLPF | FANPVLPF | 15346 | 35 |
| HLA-C*12:02 | FDNPVLPF | FANPVLPF | 12779 | 39 |
| HLA-C*12:03 | FDNPVLPF | FANPVLPF | 16585 | 52 |
| HLA-C*03:02 | FDNPVLPFNDGVY | FANPVLPFNDGVY | 12724 | 65 |
| HLA-C*02:02 | FDNPVLPF | FANPVLPF | 16957 | 120 |
| HLA-C*02:10 | FDNPVLPF | FANPVLPF | 16957 | 120 |
| HLA-B*35:01 | FDNPVLPF | FANPVLPF | 14924 | 111 |
| HLA-C*16:01 | FDNPVLPFNDGVY | FANPVLPFNDGVY | 20775 | 308 |
| HLA-C*12:02 | FDNPVLPFNDGVY | FANPVLPFNDGVY | 13459 | 202 |
| HLA-B*15:03 | FDNPVLPF | FANPVLPF | 3927 | 66 |
| HLA-A*30:01 | GTNGTKRFD | GTNGTKRFA | 11372 | 201 |
| HLA-C*02:02 | FDNPVLPFNDGVY | FANPVLPFNDGVY | 17854 | 318 |
| HLA-C*02:10 | FDNPVLPFNDGVY | FANPVLPFNDGVY | 17854 | 318 |
| HLA-C*12:03 | FDNPVLPFNDGVY | FANPVLPFNDGVY | 18712 | 334 |
| HLA-B*35:01 | FDNPVLPFNDGVY | FANPVLPFNDGVY | 11216 | 207 |
| HLA-C*03:02 | FDNPVLPFNDGVYF | FANPVLPFNDGVYF | 6446 | 128 |
| HLA-C*02:02 | FDNPVLPFNDGVYF | FANPVLPFNDGVYF | 15611 | 499 |
| HLA-C*02:10 | FDNPVLPFNDGVYF | FANPVLPFNDGVYF | 15611 | 499 |
| HLA-C*12:02 | FDNPVLPFNDGVYF | FANPVLPFNDGVYF | 11675 | 421 |
| HLA-B*15:01 | RFDNPVLPF | RFANPVLPF | 4355 | 183 |
| HLA-A*23:01 | RFDNPVLPF | RFANPVLPF | 327 | 23 |
| HLA-A*24:02 | RFDNPVLPF | RFANPVLPF | 457 | 34 |
| HLA-C*04:01 | RFDNPVLPF | RFANPVLPF | 411 | 5105 |
| HLA-C*14:02 | FDNPVLPF | FANPVLPF | 3648 | 294 |
| HLA-B*15:03 | RFDNPVLPF | RFANPVLPF | 171 | 16 |
| HLA-A*24:02 | KRFDNPVLPF | KRFANPVLPF | 2713 | 316 |
| HLA-A*23:01 | KRFDNPVLPF | KRFANPVLPF | 1220 | 146 |
| HLA-A*32:01 | RFDNPVLPF | RFANPVLPF | 1028 | 133 |
| HLA-C*14:02 | RFDNPVLPF | RFANPVLPF | 36 | 6 |
| HLA-A*30:01 | GTKRFDNPV | GTKRFANPV | 120 | 25 |
| HLA-C*03:02 | RFDNPVLPF | RFANPVLPF | 1091 | 250 |
| HLA-A*29:02 | RFDNPVLPF | RFANPVLPF | 1026 | 276 |
| HLA-A*30:01 | GTKRFDNPVL | GTKRFANPVL | 352 | 110 |
| HLA-A*68:02 | FDNPVLPFNDGV | FANPVLPFNDGV | 503 | 164 |
| HLA-A*30:02 | RFDNPVLPFNDGVY | RFANPVLPFNDGVY | 686 | 246 |
| HLA-C*14:02 | KRFDNPVLPF | KRFANPVLPF | 405 | 152 |
| HLA-C*16:01 | RFDNPVLPF | RFANPVLPF | 1270 | 485 |
| HLA-C*07:02 | RFDNPVLPF | RFANPVLPF | 1004 | 412 |
D215G
Reference REFVFKNIDGYFKIYSKHTPINLVRDLPQGFSALEPLVDLPIGINITRFQT
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated REFVFKNIDGYFKIYSKHTPINLVRGLPQGFSALEPLVDLPIGINITRFQT
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-A*02:11 | DLPQGFSALEPL | GLPQGFSALEPL | 2822 | 38 |
| HLA-A*02:01 | DLPQGFSALEPL | GLPQGFSALEPL | 5739 | 78 |
| HLA-A*02:02 | DLPQGFSALEPL | GLPQGFSALEPL | 1894 | 30 |
| HLA-A*02:11 | DLPQGFSAL | GLPQGFSAL | 1553 | 28 |
| HLA-A*02:02 | DLPQGFSAL | GLPQGFSAL | 1520 | 31 |
| HLA-B*40:02 | RDLPQGFSAL | RGLPQGFSAL | 386 | 13573 |
| HLA-A*02:11 | DLPQGFSALEPLV | GLPQGFSALEPLV | 6434 | 216 |
| HLA-A*02:01 | DLPQGFSAL | GLPQGFSAL | 7889 | 307 |
| HLA-A*02:02 | DLPQGFSALEPLV | GLPQGFSALEPLV | 6875 | 269 |
| HLA-A*02:06 | DLPQGFSALEPL | GLPQGFSALEPL | 6309 | 284 |
| HLA-A*02:06 | DLPQGFSAL | GLPQGFSAL | 3505 | 186 |
| HLA-A*68:02 | HTPINLVRDL | HTPINLVRGL | 1815 | 188 |
| HLA-C*01:02 | DLPQGFSAL | GLPQGFSAL | 196 | 46 |
| HLA-C*14:02 | DLPQGFSAL | GLPQGFSAL | 1350 | 490 |
LLA 241-243 deletion
Reference LPQGFSALEPLVDLPIGINITRFQTLLALHRSYLTPGDSSSGWTAGAAAYYVG
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated LPQGFSALEPLVDLPIGINITRFQT‑‑‑LHRSYLTPGDSSSGWTXGAAAYYVG
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-A*30:01 | ITRFQTLLA | ITRFQT | 12 | - |
| HLA-A*31:01 | RFQTLLALHR | RFQTLHR | 12 | - |
| HLA-A*02:02 | LLALHRSYL | LHRSYL | 13 | - |
| HLA-A*02:11 | LLALHRSYL | LHRSYL | 21 | - |
| HLA-B*15:03 | TLLALHRSY | TLHRSY | 28 | - |
| HLA-B*08:01 | LLALHRSYL | LHRSYL | 30 | - |
| HLA-A*02:11 | TLLALHRSYL | TLHRSYL | 33 | - |
| HLA-B*27:05 | TRFQTLLAL | TRFQTL | 35 | - |
| HLA-A*02:02 | TLLALHRSYL | TLHRSYL | 59 | - |
| HLA-B*27:05 | ITRFQTLLAL | ITRFQTL | 60 | - |
| HLA-C*06:02 | TRFQTLLAL | TRFQTL | 65 | - |
| HLA-A*29:02 | TLLALHRSY | TLHRSY | 67 | - |
| HLA-C*07:01 | TRFQTLLAL | TRFQTL | 71 | - |
| HLA-B*15:03 | TRFQTLLAL | TRFQTL | 75 | - |
| HLA-C*07:02 | TRFQTLLAL | TRFQTL | 88 | - |
| HLA-C*14:02 | RFQTLLAL | RFQTL | 91 | - |
| HLA-B*27:05 | GINITRFQTLLAL | - | 99 | - |
| HLA-C*16:01 | LALHRSYL | LHRSYL | 101 | - |
| HLA-B*15:01 | TLLALHRSY | TLHRSY | 106 | - |
| HLA-B*14:02 | TRFQTLLAL | TRFQTL | 120 | - |
| HLA-A*30:01 | ITRFQTLLAL | ITRFQTL | 126 | - |
| HLA-B*27:05 | TRFQTLLALH | TRFQTLH | 147 | - |
| HLA-B*42:01 | LLALHRSYL | LHRSYL | 170 | - |
| HLA-A*30:02 | TLLALHRSY | TLHRSY | 172 | - |
| HLA-A*02:01 | TLLALHRSYL | TLHRSYL | 174 | - |
| HLA-C*14:02 | TRFQTLLAL | TRFQTL | 203 | - |
| HLA-A*31:01 | GINITRFQTLLALHR | GINITRFQTLHR | - | 206 |
| HLA-B*27:05 | INITRFQTLLAL | - | 213 | - |
| HLA-A*30:01 | NITRFQTLLA | NITRFQT | 214 | - |
| HLA-A*30:02 | QTLLALHRSY | QTLHRSY | 214 | - |
| HLA-A*33:03 | RFQTLLALHR | RFQTLHR | 231 | - |
| HLA-B*08:01 | LALHRSYL | LHRSYL | 242 | - |
| HLA-A*31:01 | RFQTLLALH | RFQTLH | 258 | - |
| HLA-B*27:05 | NITRFQTLLAL | - | 258 | - |
| HLA-B*08:01 | TLLALHRSYL | TLHRSYL | 284 | - |
| HLA-A*02:02 | LLALHRSYLT | LHRSYLT | 306 | - |
| HLA-A*30:01 | ITRFQTLLALHRSYL | ITRFQTLHRSYL | - | 310 |
| HLA-B*15:03 | LLALHRSY | LHRSY | 313 | - |
| HLA-A*30:02 | RFQTLLALH | RFQTLH | 338 | - |
| HLA-A*02:01 | LLALHRSYL | LHRSYL | 353 | - |
| HLA-A*30:01 | INITRFQTLLA | - | 356 | - |
| HLA-C*03:02 | TLLALHRSY | TLHRSY | 362 | - |
| HLA-A*02:02 | NITRFQTLL | NITRFQT | 363 | - |
| HLA-B*15:01 | QTLLALHRSY | QTLHRSY | 363 | - |
| HLA-B*15:01 | LLALHRSY | LHRSY | 395 | - |
| HLA-C*14:02 | LLALHRSYL | LHRSYL | 407 | - |
| HLA-C*16:01 | LLALHRSYL | LHRSYL | 420 | - |
| HLA-A*31:01 | IGINITRFQTLLALHR | IGINITRFQTLHR | - | 423 |
| HLA-A*02:06 | ITRFQTLLAL | ITRFQTL | 431 | - |
| HLA-B*15:03 | ITRFQTLLAL | ITRFQTL | 451 | - |
| HLA-B*15:03 | LLALHRSYL | LHRSYL | 458 | - |
| HLA-C*03:03 | FQTLLALHRSYL | FQTLHRSYL | 10000 | 100 |
| HLA-C*03:04 | FQTLLALHRSYL | FQTLHRSYL | 10000 | 100 |
| HLA-C*14:02 | RFQTLLALHRSY | RFQTLHRSY | 1204 | 14 |
| HLA-A*31:01 | RFQTLLALHRS | RFQTLHRS | 348 | 22003 |
| HLA-C*12:03 | FQTLLALHRSYL | FQTLHRSYL | 7447 | 141 |
| HLA-C*16:01 | FQTLLALHRSYL | FQTLHRSYL | 8479 | 186 |
| HLA-C*03:02 | FQTLLALHRSYL | FQTLHRSYL | 7964 | 177 |
| HLA-C*12:02 | FQTLLALHRSYL | FQTLHRSYL | 5579 | 197 |
| HLA-A*31:01 | TRFQTLLALHR | TRFQTLHR | 145 | 3975 |
| HLA-C*14:02 | FQTLLALHRSYL | FQTLHRSYL | 4687 | 175 |
| HLA-C*07:02 | RFQTLLALHRSY | RFQTLHRSY | 10625 | 440 |
| HLA-A*30:01 | ITRFQTLLALHR | ITRFQTLHR | 3274 | 136 |
| HLA-A*68:01 | NITRFQTLLALHR | NITRFQTLHR | 652 | 30 |
| HLA-C*14:02 | TRFQTLLALHRSY | TRFQTLHRSY | 5428 | 255 |
| HLA-B*15:03 | FQTLLALHRSYL | FQTLHRSYL | 592 | 28 |
| HLA-C*17:01 | FQTLLALHRSYL | FQTLHRSYL | 8472 | 445 |
| HLA-C*02:02 | FQTLLALHRSYL | FQTLHRSYL | 8235 | 440 |
| HLA-C*02:10 | FQTLLALHRSYL | FQTLHRSYL | 8235 | 440 |
| HLA-B*15:03 | RFQTLLALHRSYL | RFQTLHRSYL | 7029 | 419 |
| HLA-C*14:02 | RFQTLLALHRSYL | RFQTLHRSYL | 1277 | 99 |
| HLA-A*33:03 | ITRFQTLLALHR | ITRFQTLHR | 656 | 60 |
| HLA-B*27:05 | TRFQTLLALHR | TRFQTLHR | 191 | 1873 |
| HLA-A*68:01 | ITRFQTLLALHR | ITRFQTLHR | 1475 | 155 |
| HLA-A*33:03 | NITRFQTLLALHR | NITRFQTLHR | 548 | 59 |
| HLA-A*30:01 | RFQTLLALHRSYL | RFQTLHRSYL | 2049 | 249 |
| HLA-A*30:01 | ITRFQTLLALHRS | ITRFQTLHRS | 3352 | 466 |
| HLA-A*31:01 | INITRFQTLLALHR | INITRFQTLHR | 2345 | 336 |
| HLA-A*29:02 | TRFQTLLALHRSY | TRFQTLHRSY | 2415 | 407 |
| HLA-A*31:01 | ITRFQTLLALHR | ITRFQTLHR | 134 | 26 |
| HLA-A*31:01 | NITRFQTLLALHR | NITRFQTLHR | 1001 | 227 |
| HLA-B*15:01 | FQTLLALHRSYL | FQTLHRSYL | 1826 | 446 |
| HLA-B*15:03 | TRFQTLLALHRSY | TRFQTLHRSY | 396 | 103 |
| HLA-B*27:05 | TRFQTLLALHRSY | TRFQTLHRSY | 511 | 146 |
| HLA-A*30:02 | TRFQTLLALHRSY | TRFQTLHRSY | 1004 | 366 |
| HLA-B*15:01 | FQTLLALHRSY | FQTLHRSY | 126 | 344 |
| HLA-A*29:02 | RFQTLLALHRSY | RFQTLHRSY | 489 | 188 |
| HLA-B*27:05 | TRFQTLLALHRSYL | TRFQTLHRSYL | 500 | 196 |
| HLA-A*02:06 | FQTLLALHRSYL | FQTLHRSYL | 579 | 257 |
| HLA-A*30:02 | RFQTLLALHRSY | RFQTLHRSY | 140 | 66 |
A260X
Reference ITRFQTLLALHRSYLTPGDSSSGWTAGAAAYYVGYLQPRTFLLKYNENGTI
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated ITRFQT‑‑‑LHRSYLTPGDSSSGWTXGAAAYYVGYLQPRTFLLKYNENGTI
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-A*02:11 | YLTPGDSSSGWTA | YLTPGDSSSGWTX | 79 | 3190 |
| HLA-A*02:02 | YLTPGDSSSGWTA | YLTPGDSSSGWTX | 106 | 2993 |
| HLA-C*16:01 | TAGAAAYYV | TXGAAAYYV | 374 | 9769 |
| HLA-A*02:01 | YLTPGDSSSGWTA | YLTPGDSSSGWTX | 262 | 6132 |
| HLA-A*68:02 | TAGAAAYYV | TXGAAAYYV | 30 | 454 |
| HLA-B*15:03 | AGAAAYYVGY | XGAAAYYVGY | 388 | 1417 |
| HLA-B*15:01 | AGAAAYYVGY | XGAAAYYVGY | 360 | 1274 |
| HLA-A*02:06 | TAGAAAYYV | TXGAAAYYV | 186 | 584 |
| HLA-A*30:02 | AGAAAYYVGY | XGAAAYYVGY | 100 | 313 |
| HLA-A*25:01 | WTAGAAAYY | WTXGAAAYY | 112 | 298 |
| HLA-A*02:11 | TAGAAAYYV | TXGAAAYYV | 107 | 284 |
| HLA-A*26:01 | DSSSGWTAGAAAYY | DSSSGWTXGAAAYY | 344 | 899 |
| HLA-A*26:01 | SSGWTAGAAAYY | SSGWTXGAAAYY | 438 | 1112 |
| HLA-A*68:02 | GWTAGAAAYYV | GWTXGAAAYYV | 215 | 537 |
| HLA-A*26:01 | GWTAGAAAYY | GWTXGAAAYY | 318 | 773 |
| HLA-A*26:01 | WTAGAAAYYVGY | WTXGAAAYYVGY | 166 | 391 |
| HLA-C*16:01 | SSSGWTAGAAAY | SSSGWTXGAAAY | 1106 | 478 |
| HLA-A*68:02 | WTAGAAAYYVGYL | WTXGAAAYYVGYL | 90 | 192 |
| HLA-C*14:02 | SGWTAGAAAY | SGWTXGAAAY | 616 | 297 |
| HLA-A*01:01 | GWTAGAAAYY | GWTXGAAAYY | 258 | 532 |
| HLA-A*02:02 | TAGAAAYYV | TXGAAAYYV | 340 | 700 |
| HLA-A*68:02 | DSSSGWTAGA | DSSSGWTXGA | 418 | 839 |
K417N
Reference FTNVYADSFVIRGDEVRQIAPGQTGKIADYNYKLPDDFTGCVIAWNSNNLD
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated FTNVYADSFVIRGDEVRQIAPGQTGNIADYNYKLPDDFTGCVIAWNSNNLD
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-A*03:01 | RQIAPGQTGK | RQIAPGQTGN | 49 | 23809 |
| HLA-A*11:01 | RQIAPGQTGK | RQIAPGQTGN | 103 | 29063 |
| HLA-A*68:01 | EVRQIAPGQTGK | EVRQIAPGQTGN | 144 | 32767 |
| HLA-A*03:02 | RQIAPGQTGK | RQIAPGQTGN | 123 | 26438 |
| HLA-A*68:01 | QIAPGQTGK | QIAPGQTGN | 136 | 27997 |
| HLA-A*11:01 | QIAPGQTGK | QIAPGQTGN | 165 | 32767 |
| HLA-A*03:02 | QIAPGQTGK | QIAPGQTGN | 278 | 32767 |
| HLA-A*03:01 | QIAPGQTGK | QIAPGQTGN | 296 | 32767 |
| HLA-A*32:01 | KIADYNYKL | NIADYNYKL | 58 | 4480 |
| HLA-A*30:01 | RQIAPGQTGK | RQIAPGQTGN | 275 | 9067 |
| HLA-A*68:02 | KIADYNYKL | NIADYNYKL | 1274 | 39 |
| HLA-A*03:02 | KIADYNYK | NIADYNYK | 476 | 6850 |
| HLA-A*02:06 | KIADYNYKL | NIADYNYKL | 20 | 101 |
| HLA-A*02:01 | KIADYNYKL | NIADYNYKL | 23 | 110 |
| HLA-C*17:01 | KIADYNYKL | NIADYNYKL | 155 | 669 |
| HLA-A*30:01 | TGKIADYNYK | TGNIADYNYK | 292 | 1164 |
| HLA-A*02:11 | KIADYNYKL | NIADYNYKL | 5 | 19 |
| HLA-A*68:01 | TGKIADYNYK | TGNIADYNYK | 957 | 343 |
| HLA-A*01:01 | QTGKIADYNY | QTGNIADYNY | 580 | 270 |
E484K
Reference SNLKPFERDISTEIYQAGSTPCNGVEGFNCYFPLQSYGFQPTNGVGYQPYR
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated SNLKPFERDISTEIYQAGSTPCNGVKGFNCYFPLQSYGFQPTYGVGYQPYR
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-B*40:02 | VEGFNCYFPL | VKGFNCYFPL | 31 | 7011 |
| HLA-B*40:01 | VEGFNCYFPL | VKGFNCYFPL | 76 | 15210 |
| HLA-A*11:01 | GSTPCNGVE | GSTPCNGVK | 32767 | 444 |
| HLA-A*68:02 | EGFNCYFPL | KGFNCYFPL | 117 | 3877 |
| HLA-A*30:01 | EGFNCYFPL | KGFNCYFPL | 5413 | 164 |
| HLA-A*32:01 | EGFNCYFPL | KGFNCYFPL | 6548 | 210 |
| HLA-A*30:02 | EGFNCYFPLQSY | KGFNCYFPLQSY | 1806 | 72 |
| HLA-B*15:01 | GVEGFNCYF | GVKGFNCYF | 3125 | 132 |
| HLA-A*31:01 | EGFNCYFPL | KGFNCYFPL | 4227 | 226 |
| HLA-B*15:03 | EGFNCYFPL | KGFNCYFPL | 2980 | 212 |
| HLA-A*02:11 | EGFNCYFPL | KGFNCYFPL | 2508 | 259 |
| HLA-B*15:03 | GVEGFNCYF | GVKGFNCYF | 1657 | 199 |
| HLA-A*02:06 | EGFNCYFPL | KGFNCYFPL | 1266 | 169 |
| HLA-B*35:01 | TPCNGVEGF | TPCNGVKGF | 319 | 1754 |
| HLA-B*15:03 | VEGFNCYFPLQSY | VKGFNCYFPLQSY | 1081 | 208 |
| HLA-B*35:01 | NGVEGFNCY | NGVKGFNCY | 346 | 1439 |
| HLA-C*16:01 | EGFNCYFPL | KGFNCYFPL | 1861 | 498 |
| HLA-B*15:03 | VEGFNCYFPL | VKGFNCYFPL | 943 | 280 |
N501Y
Reference GSTPCNGVEGFNCYFPLQSYGFQPTNGVGYQPYRVVVLSFELLHAPATVCG
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated GSTPCNGVKGFNCYFPLQSYGFQPTYGVGYQPYRVVVLSFELLHAPATVCG
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-A*29:02 | QSYGFQPTN | QSYGFQPTY | 26152 | 24 |
| HLA-A*29:02 | YFPLQSYGFQPTN | YFPLQSYGFQPTY | 19514 | 25 |
| HLA-B*15:01 | LQSYGFQPTN | LQSYGFQPTY | 6135 | 8 |
| HLA-B*35:01 | FPLQSYGFQPTN | FPLQSYGFQPTY | 8715 | 15 |
| HLA-C*03:02 | QSYGFQPTN | QSYGFQPTY | 12903 | 23 |
| HLA-B*15:03 | LQSYGFQPTN | LQSYGFQPTY | 2045 | 4 |
| HLA-B*15:03 | QSYGFQPTN | QSYGFQPTY | 6543 | 13 |
| HLA-C*16:01 | QSYGFQPTN | QSYGFQPTY | 10868 | 27 |
| HLA-B*35:01 | QSYGFQPTN | QSYGFQPTY | 24615 | 71 |
| HLA-C*12:03 | QSYGFQPTN | QSYGFQPTY | 12696 | 38 |
| HLA-B*15:03 | PLQSYGFQPTN | PLQSYGFQPTY | 22802 | 80 |
| HLA-C*14:02 | TNGVGYQPY | TYGVGYQPY | 5468 | 21 |
| HLA-A*29:02 | LQSYGFQPTN | LQSYGFQPTY | 28458 | 133 |
| HLA-C*03:02 | FPLQSYGFQPTN | FPLQSYGFQPTY | 23840 | 113 |
| HLA-C*03:02 | LQSYGFQPTN | LQSYGFQPTY | 23421 | 125 |
| HLA-A*30:02 | LQSYGFQPTN | LQSYGFQPTY | 7508 | 42 |
| HLA-A*30:02 | QSYGFQPTN | QSYGFQPTY | 5297 | 30 |
| HLA-B*15:03 | FPLQSYGFQPTN | FPLQSYGFQPTY | 20159 | 137 |
| HLA-C*03:02 | YFPLQSYGFQPTN | YFPLQSYGFQPTY | 27639 | 198 |
| HLA-A*29:02 | CYFPLQSYGFQPTN | CYFPLQSYGFQPTY | 27182 | 196 |
| HLA-C*14:02 | SYGFQPTN | SYGFQPTY | 20230 | 162 |
| HLA-A*29:02 | FPLQSYGFQPTN | FPLQSYGFQPTY | 32767 | 264 |
| HLA-B*15:01 | QSYGFQPTN | QSYGFQPTY | 21529 | 195 |
| HLA-C*14:02 | YFPLQSYGFQPTN | YFPLQSYGFQPTY | 18230 | 169 |
| HLA-B*15:03 | YFPLQSYGFQPTN | YFPLQSYGFQPTY | 22168 | 206 |
| HLA-A*11:01 | QSYGFQPTN | QSYGFQPTY | 22068 | 241 |
| HLA-C*16:01 | LQSYGFQPTN | LQSYGFQPTY | 27149 | 326 |
| HLA-C*12:02 | QSYGFQPTN | QSYGFQPTY | 21029 | 267 |
| HLA-B*15:01 | PLQSYGFQPTN | PLQSYGFQPTY | 32767 | 449 |
| HLA-B*35:01 | YFPLQSYGFQPTN | YFPLQSYGFQPTY | 28375 | 395 |
| HLA-C*12:03 | FPLQSYGFQPTN | FPLQSYGFQPTY | 27893 | 431 |
| HLA-C*16:01 | FPLQSYGFQPTN | FPLQSYGFQPTY | 27733 | 434 |
| HLA-C*02:02 | QSYGFQPTN | QSYGFQPTY | 26598 | 425 |
| HLA-C*02:10 | QSYGFQPTN | QSYGFQPTY | 26598 | 425 |
| HLA-A*30:02 | YFPLQSYGFQPTN | YFPLQSYGFQPTY | 18743 | 310 |
| HLA-B*15:03 | LQSYGFQPTNG | LQSYGFQPTYG | 6387 | 109 |
| HLA-B*53:01 | FPLQSYGFQPTN | FPLQSYGFQPTY | 8451 | 151 |
| HLA-C*12:03 | LQSYGFQPTN | LQSYGFQPTY | 26641 | 480 |
| HLA-A*32:01 | QSYGFQPTN | QSYGFQPTY | 16619 | 311 |
| HLA-A*30:02 | FPLQSYGFQPTN | FPLQSYGFQPTY | 21475 | 466 |
| HLA-C*03:02 | NGVGYQPY | YGVGYQPY | 10463 | 451 |
| HLA-B*57:01 | QSYGFQPTN | QSYGFQPTY | 4070 | 215 |
| HLA-B*58:01 | QSYGFQPTN | QSYGFQPTY | 1532 | 83 |
| HLA-A*02:11 | NGVGYQPYRV | YGVGYQPYRV | 5989 | 327 |
| HLA-B*57:01 | LQSYGFQPTN | LQSYGFQPTY | 3839 | 244 |
| HLA-B*58:01 | LQSYGFQPTN | LQSYGFQPTY | 1885 | 155 |
| HLA-B*15:03 | NGVGYQPY | YGVGYQPY | 4677 | 467 |
| HLA-A*31:01 | TNGVGYQPYR | TYGVGYQPYR | 811 | 84 |
| HLA-A*33:03 | TNGVGYQPYR | TYGVGYQPYR | 573 | 62 |
| HLA-A*29:02 | TNGVGYQPY | TYGVGYQPY | 2031 | 237 |
| HLA-A*29:02 | PTNGVGYQPY | PTYGVGYQPY | 1211 | 177 |
| HLA-A*02:11 | YGFQPTNGV | YGFQPTYGV | 307 | 49 |
| HLA-A*02:01 | YGFQPTNGV | YGFQPTYGV | 2160 | 459 |
| HLA-A*68:01 | PTNGVGYQPYR | PTYGVGYQPYR | 1073 | 242 |
| HLA-A*02:11 | LQSYGFQPTNGV | LQSYGFQPTYGV | 1446 | 396 |
| HLA-A*31:01 | PTNGVGYQPYR | PTYGVGYQPYR | 1377 | 406 |
| HLA-A*02:06 | YGFQPTNGV | YGFQPTYGV | 174 | 52 |
| HLA-A*33:03 | PTNGVGYQPYR | PTYGVGYQPYR | 1345 | 407 |
| HLA-A*02:02 | YGFQPTNGV | YGFQPTYGV | 868 | 287 |
| HLA-A*02:06 | QSYGFQPTNGV | QSYGFQPTYGV | 1121 | 407 |
| HLA-A*68:02 | YGFQPTNGV | YGFQPTYGV | 73 | 28 |
| HLA-A*68:02 | QSYGFQPTNGV | QSYGFQPTYGV | 122 | 48 |
| HLA-B*15:01 | GFQPTNGVGY | GFQPTYGVGY | 774 | 311 |
| HLA-A*29:02 | YGFQPTNGVGY | YGFQPTYGVGY | 1158 | 486 |
| HLA-A*31:01 | NGVGYQPYR | YGVGYQPYR | 438 | 185 |
| HLA-A*02:06 | LQSYGFQPTNGV | LQSYGFQPTYGV | 249 | 111 |
| HLA-A*30:02 | PTNGVGYQPY | PTYGVGYQPY | 465 | 210 |
| HLA-B*15:03 | FQPTNGVGY | FQPTYGVGY | 63 | 29 |
| HLA-A*02:02 | LQSYGFQPTNGV | LQSYGFQPTYGV | 594 | 280 |
| HLA-B*15:03 | GFQPTNGVGY | GFQPTYGVGY | 769 | 363 |
D614G
Reference PCSFGGVSVITPGTNTSNQVAVLYQDVNCTEVPVAIHADQLTPTWRVYSTG
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated PCSFGGVSVITPGTNTSNQVAVLYQGVNCTEVPVAIHADQLTPTWRVYSTG
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-C*05:01 | YQDVNCTEV | YQGVNCTEV | 82 | 8597 |
| HLA-C*08:02 | YQDVNCTEV | YQGVNCTEV | 89 | 8066 |
| HLA-A*02:11 | DVNCTEVPV | GVNCTEVPV | 2140 | 72 |
| HLA-A*02:02 | DVNCTEVPV | GVNCTEVPV | 4099 | 213 |
| HLA-A*68:02 | DVNCTEVPV | GVNCTEVPV | 36 | 659 |
| HLA-A*02:06 | DVNCTEVPV | GVNCTEVPV | 3807 | 208 |
| HLA-A*68:02 | QVAVLYQDV | QVAVLYQGV | 183 | 13 |
| HLA-A*68:02 | NQVAVLYQDV | NQVAVLYQGV | 650 | 52 |
| HLA-A*02:06 | QVAVLYQDV | QVAVLYQGV | 1312 | 148 |
| HLA-A*02:11 | QVAVLYQDV | QVAVLYQGV | 2169 | 268 |
| HLA-A*68:02 | NTSNQVAVLYQDV | NTSNQVAVLYQGV | 1858 | 231 |
| HLA-A*68:02 | TSNQVAVLYQDV | TSNQVAVLYQGV | 3281 | 457 |
| HLA-A*02:02 | QVAVLYQDV | QVAVLYQGV | 1107 | 188 |
| HLA-A*02:01 | YQDVNCTEV | YQGVNCTEV | 75 | 315 |
| HLA-A*02:06 | YQDVNCTEV | YQGVNCTEV | 17 | 57 |
| HLA-A*02:11 | VLYQDVNCT | VLYQGVNCT | 224 | 74 |
| HLA-A*02:11 | YQDVNCTEV | YQGVNCTEV | 31 | 93 |
| HLA-A*02:11 | VLYQDVNCTEVP | VLYQGVNCTEVP | 1000 | 402 |
| HLA-A*02:02 | NQVAVLYQDV | NQVAVLYQGV | 269 | 116 |
| HLA-A*02:11 | AVLYQDVNCTEV | AVLYQGVNCTEV | 500 | 238 |
| HLA-A*02:01 | NQVAVLYQDV | NQVAVLYQGV | 558 | 267 |
| HLA-A*02:06 | NQVAVLYQDV | NQVAVLYQGV | 36 | 18 |
A701V
Reference TQTNSPRRARSVASQSIIAYTMSLGAENSVAYSNNSIAIPTNFTISVTTEI
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated TQTNSPRRARSVASQSIIAYTMSLGVENSVAYSNNSIAIPTNFTISVTTEI
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-C*15:02 | IAYTMSLGA | IAYTMSLGV | 1895 | 45 |
| HLA-A*02:11 | SIIAYTMSLGA | SIIAYTMSLGV | 954 | 32 |
| HLA-A*02:11 | IIAYTMSLGA | IIAYTMSLGV | 834 | 30 |
| HLA-A*02:11 | IAYTMSLGA | IAYTMSLGV | 2062 | 76 |
| HLA-C*17:01 | IAYTMSLGA | IAYTMSLGV | 4933 | 248 |
| HLA-C*12:03 | IAYTMSLGA | IAYTMSLGV | 289 | 16 |
| HLA-A*02:01 | SIIAYTMSLGA | SIIAYTMSLGV | 1860 | 107 |
| HLA-A*02:01 | IIAYTMSLGA | IIAYTMSLGV | 1312 | 91 |
| HLA-A*02:06 | IAYTMSLGA | IAYTMSLGV | 739 | 57 |
| HLA-C*16:01 | IAYTMSLGA | IAYTMSLGV | 553 | 43 |
| HLA-C*03:03 | IAYTMSLGA | IAYTMSLGV | 1287 | 101 |
| HLA-C*03:04 | IAYTMSLGA | IAYTMSLGV | 1287 | 101 |
| HLA-A*02:01 | IAYTMSLGA | IAYTMSLGV | 3643 | 300 |
| HLA-A*68:02 | IIAYTMSLGA | IIAYTMSLGV | 1653 | 144 |
| HLA-B*51:01 | IAYTMSLGA | IAYTMSLGV | 5547 | 488 |
| HLA-A*02:06 | SIIAYTMSLGA | SIIAYTMSLGV | 706 | 64 |
| HLA-B*44:02 | AENSVAYSNNSI | VENSVAYSNNSI | 154 | 1676 |
| HLA-C*02:02 | IAYTMSLGA | IAYTMSLGV | 3671 | 343 |
| HLA-C*02:10 | IAYTMSLGA | IAYTMSLGV | 3671 | 343 |
| HLA-A*02:02 | IIAYTMSLGA | IIAYTMSLGV | 401 | 38 |
| HLA-A*68:02 | SIIAYTMSLGA | SIIAYTMSLGV | 3371 | 320 |
| HLA-B*45:01 | AENSVAYSNNSIA | VENSVAYSNNSIA | 83 | 849 |
| HLA-A*02:02 | SIIAYTMSLGA | SIIAYTMSLGV | 1180 | 122 |
| HLA-A*02:06 | IIAYTMSLGA | IIAYTMSLGV | 819 | 87 |
| HLA-A*68:02 | IAYTMSLGA | IAYTMSLGV | 2063 | 226 |
| HLA-B*44:02 | AENSVAYSN | VENSVAYSN | 233 | 1867 |
| HLA-B*44:03 | AENSVAYSNNSI | VENSVAYSNNSI | 367 | 2921 |
| HLA-A*02:06 | SQSIIAYTMSLGA | SQSIIAYTMSLGV | 2545 | 336 |
| HLA-A*02:02 | IAYTMSLGA | IAYTMSLGV | 2768 | 367 |
| HLA-C*12:02 | IAYTMSLGA | IAYTMSLGV | 1418 | 192 |
| HLA-B*44:03 | AENSVAYSN | VENSVAYSN | 237 | 1574 |
| HLA-B*45:01 | AENSVAYSN | VENSVAYSN | 392 | 2480 |
| HLA-C*03:02 | IAYTMSLGA | IAYTMSLGV | 530 | 93 |
| HLA-A*68:02 | MSLGAENSV | MSLGVENSV | 95 | 32 |
| HLA-A*02:11 | TMSLGAENSV | TMSLGVENSV | 110 | 41 |
| HLA-A*02:11 | MSLGAENSV | MSLGVENSV | 513 | 192 |
| HLA-A*02:02 | TMSLGAENSV | TMSLGVENSV | 74 | 30 |
| HLA-A*02:01 | TMSLGAENSV | TMSLGVENSV | 455 | 186 |
| HLA-B*40:02 | AENSVAYSNNSI | VENSVAYSNNSI | 463 | 1070 |
| HLA-A*02:06 | MSLGAENSV | MSLGVENSV | 188 | 92 |
| HLA-A*30:02 | LGAENSVAY | LGVENSVAY | 367 | 745 |
N protein
T205I
Reference SQASSRSSSRSRNSSRNSTPGSSRGTSPARMAGNGGDAALALLLLDRLNQL
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated SQASSRSSSRSRNSSRNSTPGSSRGISPARMAGNGGDAALALLLLDRLNQL
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-C*01:02 | STPGSSRGT | STPGSSRGI | 10810 | 468 |
| HLA-A*30:01 | GTSPARMAG | GISPARMAG | 493 | 1901 |
| HLA-A*68:01 | SSRGTSPAR | SSRGISPAR | 186 | 61 |
| HLA-A*68:01 | STPGSSRGTSPAR | STPGSSRGISPAR | 860 | 288 |
| HLA-A*30:01 | RGTSPARMA | RGISPARMA | 1160 | 422 |
| HLA-A*68:01 | GSSRGTSPAR | GSSRGISPAR | 615 | 248 |
| HLA-A*31:01 | GSSRGTSPAR | GSSRGISPAR | 262 | 111 |
| HLA-A*33:03 | SSRGTSPAR | SSRGISPAR | 106 | 46 |
| HLA-A*33:03 | STPGSSRGTSPAR | STPGSSRGISPAR | 968 | 473 |
E protein

P71L
Reference IVNVSLVKPSFYVYSRVKNLNSSRVPDLLV
||||||||||||||||||||||||| ||||
Mutated IVNVSLVKPSFYVYSRVKNLNSSRVLDLLV
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-B*15:03 | VKNLNSSRVP | VKNLNSSRVL | 13920 | 189 |
| HLA-B*15:03 | KNLNSSRVP | KNLNSSRVL | 17021 | 397 |
| HLA-A*30:01 | RVKNLNSSRVP | RVKNLNSSRVL | 1083 | 219 |
| HLA-A*68:02 | NSSRVPDLL | NSSRVLDLL | 1072 | 280 |
| HLA-A*02:02 | NLNSSRVPDLL | NLNSSRVLDLL | 657 | 201 |
| HLA-A*68:02 | NSSRVPDLLV | NSSRVLDLLV | 608 | 206 |
| HLA-A*02:02 | NLNSSRVPDLLV | NLNSSRVLDLLV | 740 | 327 |
NS3 protein
Q57H
Reference TATIPIQASLPFGWLIVGVALLAVFQSASKIITLKKRWQLALSKGVHFVCN
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated TATIPIQASLPFGWLIVGVALLAVFHSASKIITLKKRWQLALSKGVHFVCN
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-B*15:01 | FQSASKIITL | FHSASKIITL | 156 | 6196 |
| HLA-A*02:06 | FQSASKIITL | FHSASKIITL | 43 | 1441 |
| HLA-A*02:01 | FQSASKIITL | FHSASKIITL | 205 | 5629 |
| HLA-A*02:11 | FQSASKIITL | FHSASKIITL | 79 | 2010 |
| HLA-A*02:02 | FQSASKIITL | FHSASKIITL | 38 | 957 |
| HLA-B*38:01 | FQSASKIITL | FHSASKIITL | 2071 | 94 |
| HLA-B*15:03 | FQSASKIITL | FHSASKIITL | 88 | 851 |
| HLA-C*03:03 | QSASKIITL | HSASKIITL | 285 | 37 |
| HLA-C*03:04 | QSASKIITL | HSASKIITL | 285 | 37 |
| HLA-C*03:02 | QSASKIITL | HSASKIITL | 724 | 94 |
| HLA-C*12:03 | QSASKIITL | HSASKIITL | 250 | 34 |
| HLA-C*12:02 | QSASKIITL | HSASKIITL | 1730 | 284 |
| HLA-C*02:02 | QSASKIITL | HSASKIITL | 2653 | 440 |
| HLA-C*02:10 | QSASKIITL | HSASKIITL | 2653 | 440 |
| HLA-A*68:02 | QSASKIITL | HSASKIITL | 1085 | 232 |
| HLA-C*17:01 | QSASKIITL | HSASKIITL | 453 | 102 |
| HLA-A*30:01 | QSASKIITLK | HSASKIITLK | 727 | 164 |
| HLA-A*30:01 | LAVFQSASK | LAVFHSASK | 1169 | 264 |
| HLA-C*15:02 | QSASKIITL | HSASKIITL | 222 | 51 |
| HLA-C*16:01 | QSASKIITL | HSASKIITL | 52 | 12 |
| HLA-A*68:01 | QSASKIITLKK | HSASKIITLKK | 1356 | 322 |
| HLA-A*30:01 | QSASKIITLKK | HSASKIITLKK | 1813 | 486 |
| HLA-A*68:01 | QSASKIITLKKR | HSASKIITLKKR | 683 | 195 |
| HLA-A*68:01 | QSASKIITLK | HSASKIITLK | 35 | 10 |
| HLA-A*02:11 | LLAVFQSA | LLAVFHSA | 1121 | 356 |
| HLA-A*31:01 | QSASKIITLKKR | HSASKIITLKKR | 968 | 344 |
| HLA-A*03:01 | QSASKIITLK | HSASKIITLK | 214 | 83 |
| HLA-A*03:01 | AVFQSASKIITLKK | AVFHSASKIITLKK | 227 | 90 |
| HLA-A*02:02 | LLAVFQSASKII | LLAVFHSASKII | 298 | 727 |
| HLA-A*03:01 | AVFQSASK | AVFHSASK | 795 | 335 |
| HLA-A*03:01 | AVFQSASKIITLK | AVFHSASKIITLK | 74 | 32 |
| HLA-A*03:02 | AVFQSASKIITLKK | AVFHSASKIITLKK | 619 | 272 |
| HLA-A*02:11 | GVALLAVFQSA | GVALLAVFHSA | 895 | 401 |
| HLA-A*68:01 | AVFQSASKIITLK | AVFHSASKIITLK | 808 | 368 |
| HLA-A*03:02 | AVFQSASKIITLK | AVFHSASKIITLK | 170 | 82 |
| HLA-A*02:11 | VALLAVFQSA | VALLAVFHSA | 173 | 85 |
| HLA-A*02:11 | ALLAVFQSAS | ALLAVFHSAS | 691 | 340 |
| HLA-A*02:01 | ALLAVFQSA | ALLAVFHSA | 42 | 21 |
| HLA-B*58:01 | QSASKIITLKKRW | HSASKIITLKKRW | 142 | 71 |
S171L
Reference FLCWHTNCYDYCIPYNSVTSSIVITSGDGTTSPISEHDYQIGGYTEKWESG
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated FLCWHTNCYDYCIPYNSVTSSIVITLGDGTTSPISEHDYQIGGYTEKWESG
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-C*17:01 | VTSSIVITS | VTSSIVITL | 13914 | 69 |
| HLA-C*03:03 | VTSSIVITS | VTSSIVITL | 19726 | 117 |
| HLA-C*03:04 | VTSSIVITS | VTSSIVITL | 19726 | 117 |
| HLA-A*32:01 | VTSSIVITS | VTSSIVITL | 13457 | 98 |
| HLA-C*16:01 | VTSSIVITS | VTSSIVITL | 4293 | 36 |
| HLA-A*02:11 | TSGDGTTSPI | TLGDGTTSPI | 10425 | 92 |
| HLA-A*02:11 | VTSSIVITS | VTSSIVITL | 8785 | 89 |
| HLA-A*02:02 | TSGDGTTSPI | TLGDGTTSPI | 8938 | 95 |
| HLA-C*12:03 | VTSSIVITS | VTSSIVITL | 4793 | 51 |
| HLA-C*15:02 | VTSSIVITS | VTSSIVITL | 7397 | 89 |
| HLA-C*03:02 | VTSSIVITS | VTSSIVITL | 9955 | 175 |
| HLA-A*02:02 | VTSSIVITS | VTSSIVITL | 5314 | 103 |
| HLA-C*16:01 | TSSIVITS | TSSIVITL | 18534 | 362 |
| HLA-B*42:01 | IPYNSVTSSIVITS | IPYNSVTSSIVITL | 14524 | 360 |
| HLA-A*02:11 | SVTSSIVITS | SVTSSIVITL | 12237 | 410 |
| HLA-A*02:02 | YNSVTSSIVITS | YNSVTSSIVITL | 8810 | 301 |
| HLA-A*02:02 | SVTSSIVITS | SVTSSIVITL | 5294 | 197 |
| HLA-C*12:02 | VTSSIVITS | VTSSIVITL | 9810 | 369 |
| HLA-B*58:01 | VTSSIVITS | VTSSIVITL | 2482 | 96 |
| HLA-A*02:06 | VTSSIVITS | VTSSIVITL | 4179 | 183 |
| HLA-A*68:02 | SVTSSIVITS | SVTSSIVITL | 1418 | 92 |
| HLA-A*68:02 | YNSVTSSIVITS | YNSVTSSIVITL | 2711 | 182 |
| HLA-A*68:02 | VTSSIVITS | VTSSIVITL | 911 | 63 |
| HLA-A*68:02 | NSVTSSIVITS | NSVTSSIVITL | 5500 | 388 |
| HLA-C*08:02 | SGDGTTSPI | LGDGTTSPI | 222 | 110 |
NS9c protein
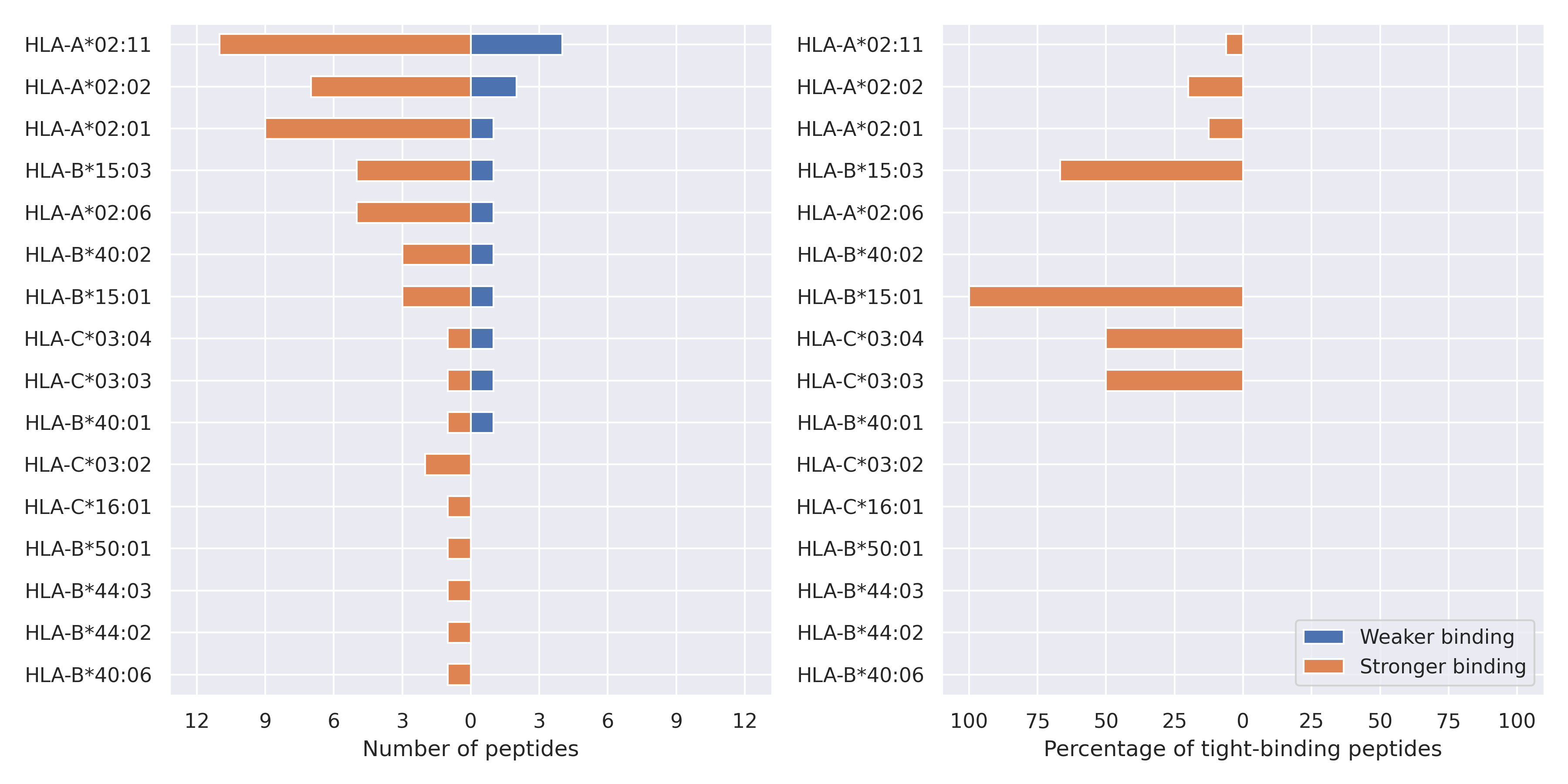
L52F
Reference VKPLLVPHHVVATVQEIQLQAAVGELLLLEWLAMAVMLLLLCCCLTD
||||||||||||||||||||||||| |||||||||||||||||||||
Mutated VKPLLVPHHVVATVQEIQLQAAVGEFLLLEWLAMAVMLLLLCCCLTD
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-A*02:11 | QLQAAVGEL | QLQAAVGEF | 120 | 5031 |
| HLA-A*02:11 | GELLLLEWLA | GEFLLLEWLA | 267 | 9767 |
| HLA-B*15:01 | QLQAAVGEL | QLQAAVGEF | 2048 | 65 |
| HLA-A*02:01 | GELLLLEWLA | GEFLLLEWLA | 409 | 12481 |
| HLA-A*02:02 | QLQAAVGEL | QLQAAVGEF | 92 | 2789 |
| HLA-B*15:03 | QLQAAVGEL | QLQAAVGEF | 2231 | 81 |
| HLA-A*02:02 | IQLQAAVGEL | IQLQAAVGEF | 170 | 4413 |
| HLA-A*02:11 | ELLLLEWLA | EFLLLEWLA | 345 | 8719 |
| HLA-A*02:11 | IQLQAAVGEL | IQLQAAVGEF | 338 | 7506 |
| HLA-B*15:01 | IQLQAAVGEL | IQLQAAVGEF | 467 | 24 |
| HLA-B*15:03 | IQLQAAVGEL | IQLQAAVGEF | 168 | 10 |
| HLA-B*15:03 | LQAAVGEL | LQAAVGEF | 295 | 18 |
| HLA-B*15:03 | VQEIQLQAAVGEL | VQEIQLQAAVGEF | 6575 | 430 |
| HLA-A*02:11 | LLLLEWLAM | FLLLEWLAM | 283 | 20 |
| HLA-B*40:01 | QEIQLQAAVGEL | QEIQLQAAVGEF | 201 | 2708 |
| HLA-B*15:01 | LQAAVGEL | LQAAVGEF | 1712 | 138 |
| HLA-A*02:01 | LLLLEWLAM | FLLLEWLAM | 573 | 54 |
| HLA-A*02:11 | LLLLEWLA | FLLLEWLA | 844 | 82 |
| HLA-A*02:02 | LLLLEWLAM | FLLLEWLAM | 1379 | 150 |
| HLA-B*44:03 | QEIQLQAAVGEL | QEIQLQAAVGEF | 1386 | 164 |
| HLA-A*02:06 | LLLLEWLAM | FLLLEWLAM | 763 | 94 |
| HLA-A*02:01 | LLLLEWLA | FLLLEWLA | 1287 | 163 |
| HLA-B*44:02 | QEIQLQAAVGEL | QEIQLQAAVGEF | 1403 | 179 |
| HLA-A*02:02 | LLLLEWLA | FLLLEWLA | 2866 | 389 |
| HLA-C*03:02 | LLLLEWLAM | FLLLEWLAM | 2566 | 358 |
| HLA-A*02:06 | IQLQAAVGEL | IQLQAAVGEF | 280 | 1893 |
| HLA-A*02:06 | LLLLEWLA | FLLLEWLA | 2803 | 489 |
| HLA-A*02:02 | LLLLEWLAMA | FLLLEWLAMA | 67 | 13 |
| HLA-A*02:01 | ELLLLEWLAMA | EFLLLEWLAMA | 1276 | 266 |
| HLA-A*02:11 | QLQAAVGELLL | QLQAAVGEFLL | 733 | 157 |
| HLA-B*40:02 | QEIQLQAAVGEL | QEIQLQAAVGEF | 334 | 1488 |
| HLA-A*02:06 | LLLLEWLAMA | FLLLEWLAMA | 49 | 11 |
| HLA-A*02:11 | LLLLEWLAMAVM | FLLLEWLAMAVM | 926 | 212 |
| HLA-A*02:02 | LLLLEWLAMAVMLL | FLLLEWLAMAVMLL | 714 | 164 |
| HLA-A*02:01 | LLLLEWLAMAVML | FLLLEWLAMAVML | 1353 | 320 |
| HLA-A*02:11 | LLLLEWLAMAVML | FLLLEWLAMAVML | 385 | 97 |
| HLA-A*02:11 | LQAAVGELLL | LQAAVGEFLL | 1331 | 346 |
| HLA-A*02:01 | LLLLEWLAMA | FLLLEWLAMA | 23 | 6 |
| HLA-A*02:11 | ELLLLEWLAMA | EFLLLEWLAMA | 299 | 79 |
| HLA-A*02:11 | LLLLEWLAMAVMLL | FLLLEWLAMAVMLL | 544 | 146 |
| HLA-A*02:01 | LLLLEWLAMAVMLL | FLLLEWLAMAVMLL | 1361 | 373 |
| HLA-B*40:01 | GELLLLEWLAM | GEFLLLEWLAM | 791 | 222 |
| HLA-B*15:03 | AAVGELLLL | AAVGEFLLL | 1475 | 426 |
| HLA-A*02:01 | QLQAAVGELLL | QLQAAVGEFLL | 1401 | 441 |
| HLA-A*02:02 | LLLLEWLAMAV | FLLLEWLAMAV | 453 | 147 |
| HLA-C*03:03 | QAAVGELLL | QAAVGEFLL | 283 | 829 |
| HLA-C*03:04 | QAAVGELLL | QAAVGEFLL | 283 | 829 |
| HLA-A*02:02 | QLQAAVGELLL | QLQAAVGEFLL | 530 | 186 |
| HLA-B*50:01 | GELLLLEWLA | GEFLLLEWLA | 1298 | 484 |
| HLA-A*02:11 | LLLLEWLAMA | FLLLEWLAMA | 8 | 3 |
| HLA-A*02:01 | LQAAVGELLL | LQAAVGEFLL | 917 | 347 |
| HLA-C*03:03 | AAVGELLLL | AAVGEFLLL | 107 | 41 |
| HLA-C*03:04 | AAVGELLLL | AAVGEFLLL | 107 | 41 |
| HLA-A*02:06 | LQAAVGELLL | LQAAVGEFLL | 162 | 63 |
| HLA-B*40:06 | GELLLLEWLA | GEFLLLEWLA | 268 | 106 |
| HLA-B*40:02 | GELLLLEWLAM | GEFLLLEWLAM | 201 | 83 |
| HLA-A*02:02 | LQAAVGELLL | LQAAVGEFLL | 100 | 42 |
| HLA-C*03:02 | AAVGELLLL | AAVGEFLLL | 269 | 116 |
| HLA-A*02:01 | LLLLEWLAMAV | FLLLEWLAMAV | 97 | 44 |
| HLA-A*02:11 | QLQAAVGELL | QLQAAVGEFL | 303 | 139 |
| HLA-B*15:01 | LQAAVGELLL | LQAAVGEFLL | 109 | 237 |
| HLA-B*15:03 | LQAAVGELLL | LQAAVGEFLL | 98 | 210 |
| HLA-A*02:11 | LQAAVGELL | LQAAVGEFL | 386 | 183 |
| HLA-B*40:02 | GELLLLEWLAMAV | GEFLLLEWLAMAV | 815 | 391 |
| HLA-B*40:02 | GELLLLEWLA | GEFLLLEWLA | 475 | 231 |
| HLA-A*02:06 | IQLQAAVGELLL | IQLQAAVGEFLL | 626 | 310 |
| HLA-C*16:01 | AAVGELLLL | AAVGEFLLL | 205 | 102 |
NSP2 protein
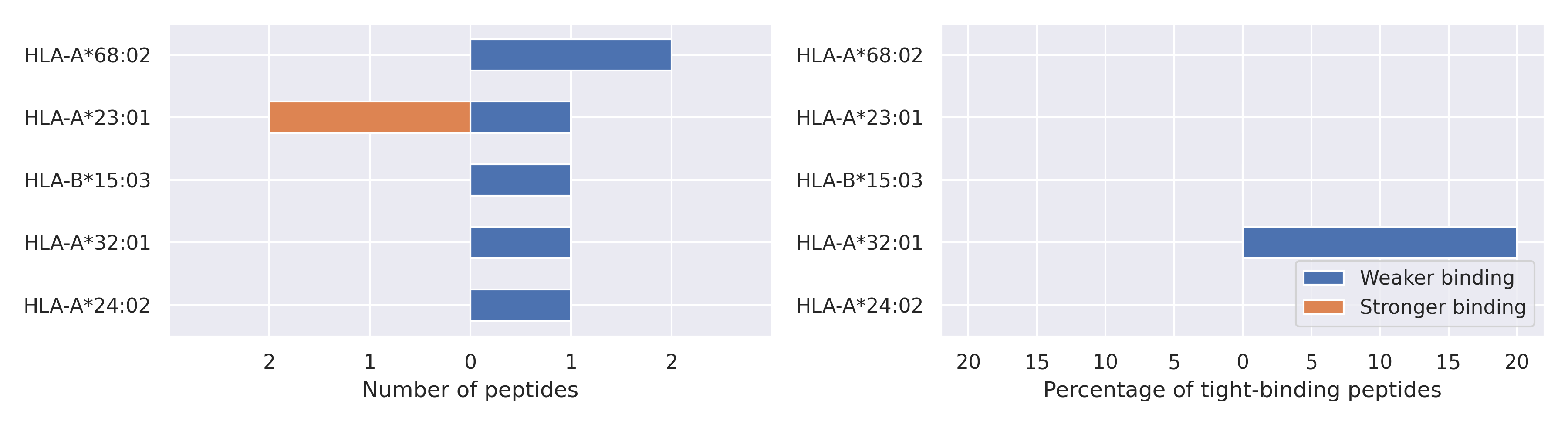
T85I
Reference WYTERSEKSYELQTPFEIKLAKKFDTFNGECPNFVFPLNSIIKTIQPRVEK
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated WYTERSEKSYELQTPFEIKLAKKFDIFNGECPNFVFPLNSIIKTIQPRVEK
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-A*68:02 | DTFNGECPNFVFPL | DIFNGECPNFVFPL | 203 | 1683 |
| HLA-A*68:02 | DTFNGECPNFV | DIFNGECPNFV | 60 | 311 |
| HLA-A*32:01 | KLAKKFDTF | KLAKKFDIF | 44 | 159 |
| HLA-A*24:02 | KLAKKFDTF | KLAKKFDIF | 237 | 578 |
| HLA-B*15:03 | KKFDTFNGECPNF | KKFDIFNGECPNF | 226 | 542 |
| HLA-A*23:01 | TFNGECPNF | IFNGECPNF | 517 | 219 |
| HLA-A*23:01 | TFNGECPNFVF | IFNGECPNFVF | 425 | 191 |
| HLA-A*23:01 | KLAKKFDTF | KLAKKFDIF | 284 | 624 |
NSP3 protein
K837N
Reference EAFEYYHTTDPSFLGRYMSALNHTKKWKYPQVNGLTSIKWADNNCYLATAL
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated EAFEYYHTTDPSFLGRYMSALNHTKNWKYPQVNGLTSIKWADNNCYLATAL
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-A*68:01 | MSALNHTKK | MSALNHTKN | 25 | 16109 |
| HLA-A*11:01 | MSALNHTKK | MSALNHTKN | 55 | 28173 |
| HLA-A*03:02 | MSALNHTKK | MSALNHTKN | 225 | 30854 |
| HLA-A*03:01 | MSALNHTKK | MSALNHTKN | 352 | 32755 |
| HLA-A*30:01 | MSALNHTKK | MSALNHTKN | 101 | 7035 |
| HLA-A*68:01 | YMSALNHTKK | YMSALNHTKN | 159 | 3833 |
| HLA-A*11:01 | YMSALNHTKK | YMSALNHTKN | 170 | 3650 |
| HLA-A*30:01 | KWKYPQVNG | NWKYPQVNG | 220 | 4236 |
| HLA-A*03:01 | YMSALNHTKK | YMSALNHTKN | 135 | 2562 |
| HLA-A*30:01 | KWKYPQVNGL | NWKYPQVNGL | 419 | 7410 |
| HLA-A*03:02 | YMSALNHTKK | YMSALNHTKN | 52 | 729 |
| HLA-A*31:01 | RYMSALNHTKK | RYMSALNHTKN | 106 | 777 |
| HLA-A*03:02 | SALNHTKKWK | SALNHTKNWK | 1062 | 167 |
| HLA-A*30:01 | RYMSALNHTKK | RYMSALNHTKN | 171 | 1048 |
| HLA-A*11:01 | SALNHTKKWK | SALNHTKNWK | 673 | 117 |
| HLA-A*11:01 | ALNHTKKWK | ALNHTKNWK | 1758 | 323 |
| HLA-A*03:02 | ALNHTKKWK | ALNHTKNWK | 266 | 53 |
| HLA-A*68:02 | HTKKWKYPQV | HTKNWKYPQV | 1273 | 380 |
| HLA-A*30:02 | ALNHTKKWKY | ALNHTKNWKY | 693 | 278 |
| HLA-A*03:01 | SALNHTKKWK | SALNHTKNWK | 506 | 204 |
| HLA-B*27:05 | GRYMSALNHTKK | GRYMSALNHTKN | 140 | 336 |
| HLA-A*03:01 | ALNHTKKWK | ALNHTKNWK | 132 | 60 |
| HLA-A*30:01 | SALNHTKKWK | SALNHTKNWK | 535 | 251 |
NSP6 protein
SGF 106-108 deletion
Reference FNMVYMPASWVMRIMTWLDMVDTSLSGFKLKDCVMYASAVVLLILMTARTVYD
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated FNMVYMPASWVMRIMTWLDMVDTSL‑‑‑KLKDCVMYASAVVLLILMTARTVYD
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-A*02:02 | FKLKDCVMYA | - | 8 | - |
| HLA-A*02:11 | FKLKDCVMYA | - | 12 | - |
| HLA-A*68:01 | MVDTSLSGFK | MVDTSLK | 13 | - |
| HLA-B*15:03 | FKLKDCVMY | - | 15 | - |
| HLA-A*11:01 | MVDTSLSGFK | MVDTSLK | 21 | - |
| HLA-A*11:01 | TSLSGFKLK | TSLKLK | 22 | - |
| HLA-A*03:01 | RIMTWLDMVDTSLSGFK | RIMTWLDMVDTSLK | - | 25 |
| HLA-A*02:01 | FKLKDCVMYA | - | 27 | - |
| HLA-A*68:01 | DTSLSGFKLK | DTSLKLK | 33 | - |
| HLA-A*03:02 | MVDTSLSGFK | MVDTSLK | 35 | - |
| HLA-A*68:01 | MTWLDMVDTSLSGFK | MTWLDMVDTSLK | - | 37 |
| HLA-A*03:01 | MVDTSLSGFK | MVDTSLK | 54 | - |
| HLA-A*03:02 | RIMTWLDMVDTSLSGFK | RIMTWLDMVDTSLK | - | 59 |
| HLA-C*05:01 | MVDTSLSGF | MVDTSL | 64 | - |
| HLA-A*11:01 | MTWLDMVDTSLSGFK | MTWLDMVDTSLK | - | 71 |
| HLA-A*02:06 | FKLKDCVMYA | - | 90 | - |
| HLA-A*30:01 | TSLSGFKLK | TSLKLK | 107 | - |
| HLA-A*02:02 | SGFKLKDCVMYA | - | 126 | - |
| HLA-A*11:01 | RIMTWLDMVDTSLSGFK | RIMTWLDMVDTSLK | - | 128 |
| HLA-A*03:02 | MTWLDMVDTSLSGFK | MTWLDMVDTSLK | - | 147 |
| HLA-C*08:02 | MVDTSLSGF | MVDTSL | 159 | - |
| HLA-A*03:02 | IMTWLDMVDTSLSGFK | IMTWLDMVDTSLK | - | 168 |
| HLA-A*02:11 | SGFKLKDCVMYA | - | 200 | - |
| HLA-A*02:02 | MTWLDMVDTSLSGFKL | MTWLDMVDTSLKL | - | 202 |
| HLA-A*01:01 | MVDTSLSGFKLKDCVMY | MVDTSLKLKDCVMY | - | 207 |
| HLA-C*03:02 | MVDTSLSGF | MVDTSL | 211 | - |
| HLA-A*02:02 | GFKLKDCVMYA | - | 239 | - |
| HLA-A*02:02 | TSLSGFKLKDCVMYA | TSLKLKDCVMYA | - | 244 |
| HLA-A*03:01 | TSLSGFKLK | TSLKLK | 244 | - |
| HLA-B*15:03 | MVDTSLSGF | MVDTSL | 276 | - |
| HLA-A*68:01 | TSLSGFKLK | TSLKLK | 286 | - |
| HLA-A*03:02 | TSLSGFKLK | TSLKLK | 289 | - |
| HLA-A*03:01 | IMTWLDMVDTSLSGFK | IMTWLDMVDTSLK | - | 309 |
| HLA-A*68:02 | DTSLSGFKL | DTSLKL | 313 | - |
| HLA-A*02:11 | MTWLDMVDTSLSGFKL | MTWLDMVDTSLKL | - | 346 |
| HLA-B*15:01 | DMVDTSLSGF | DMVDTSL | 362 | - |
| HLA-A*23:01 | TWLDMVDTSLSGF | - | 387 | - |
| HLA-A*02:11 | GFKLKDCVMYA | - | 394 | - |
| HLA-A*03:01 | MTWLDMVDTSLSGFK | MTWLDMVDTSLK | - | 415 |
| HLA-A*26:01 | DMVDTSLSGF | DMVDTSL | 417 | - |
| HLA-B*08:01 | MVDTSLSGFKLKDCVM | MVDTSLKLKDCVM | - | 426 |
| HLA-A*02:02 | WLDMVDTSLS | - | 441 | - |
| HLA-A*02:11 | FKLKDCVMYASAVV | - | 481 | - |
| HLA-A*11:01 | DTSLSGFKLK | DTSLKLK | 488 | - |
| HLA-B*35:01 | MVDTSLSGF | MVDTSL | 490 | - |
| HLA-A*31:01 | RIMTWLDMVDTSLSGFK | RIMTWLDMVDTSLK | - | 493 |
| HLA-B*15:03 | LSGFKLKDCVMY | LKLKDCVMY | 3466 | 22 |
| HLA-A*02:11 | LDMVDTSLSGFKL | LDMVDTSLKL | 16396 | 166 |
| HLA-A*02:02 | LDMVDTSLSGFKL | LDMVDTSLKL | 9971 | 106 |
| HLA-A*02:11 | SLSGFKLKDCV | SLKLKDCV | 122 | 9934 |
| HLA-A*02:01 | LDMVDTSLSGFKL | LDMVDTSLKL | 20579 | 311 |
| HLA-B*08:01 | SLSGFKLKDCVM | SLKLKDCVM | 929 | 36 |
| HLA-A*02:02 | SLSGFKLKDCV | SLKLKDCV | 408 | 10216 |
| HLA-C*05:01 | WLDMVDTSLSGFKL | WLDMVDTSLKL | 8375 | 417 |
| HLA-B*15:01 | SLSGFKLKDCVMY | SLKLKDCVMY | 2952 | 158 |
| HLA-B*08:01 | TSLSGFKLKDCVM | TSLKLKDCVM | 4628 | 289 |
| HLA-B*15:01 | SLSGFKLKDCVM | SLKLKDCVM | 2654 | 197 |
| HLA-A*68:01 | DMVDTSLSGFK | DMVDTSLK | 366 | 4919 |
| HLA-A*02:11 | WLDMVDTSLSGFKL | WLDMVDTSLKL | 1342 | 108 |
| HLA-C*08:02 | MVDTSLSGFKL | MVDTSLKL | 2134 | 174 |
| HLA-B*15:03 | SLSGFKLKDCVM | SLKLKDCVM | 3634 | 297 |
| HLA-A*02:01 | WLDMVDTSLSGFKL | WLDMVDTSLKL | 2214 | 195 |
| HLA-A*03:02 | WLDMVDTSLSGFK | WLDMVDTSLK | 1756 | 156 |
| HLA-A*02:02 | LSGFKLKDCVMYA | LKLKDCVMYA | 305 | 28 |
| HLA-A*02:01 | LSGFKLKDCVMYA | LKLKDCVMYA | 1788 | 165 |
| HLA-A*02:11 | LSGFKLKDCVMYA | LKLKDCVMYA | 578 | 54 |
| HLA-B*15:03 | SLSGFKLKDCVMY | SLKLKDCVMY | 3626 | 349 |
| HLA-A*03:01 | WLDMVDTSLSGFK | WLDMVDTSLK | 2900 | 287 |
| HLA-C*05:01 | MVDTSLSGFKL | MVDTSLKL | 1125 | 124 |
| HLA-A*11:01 | WLDMVDTSLSGFK | WLDMVDTSLK | 4020 | 492 |
| HLA-A*02:02 | WLDMVDTSLSGFKL | WLDMVDTSLKL | 788 | 116 |
| HLA-A*68:01 | DMVDTSLSGFKLK | DMVDTSLKLK | 1679 | 251 |
| HLA-A*02:01 | SLSGFKLKDCVMYA | SLKLKDCVMYA | 203 | 853 |
| HLA-A*02:11 | SLSGFKLKDCVMYA | SLKLKDCVMYA | 64 | 223 |
| HLA-A*02:02 | SLSGFKLKDCVMYA | SLKLKDCVMYA | 55 | 171 |
| HLA-A*29:02 | SLSGFKLKDCVMY | SLKLKDCVMY | 404 | 1192 |
| HLA-A*11:01 | MVDTSLSGFKLK | MVDTSLKLK | 143 | 50 |
| HLA-A*03:02 | MVDTSLSGFKLK | MVDTSLKLK | 312 | 127 |
| HLA-A*68:01 | MVDTSLSGFKLK | MVDTSLKLK | 602 | 246 |
| HLA-A*03:01 | MVDTSLSGFKLK | MVDTSLKLK | 441 | 190 |
NSP12 protein
P323L
Reference CVNCLDDRCILHCANFNVLFSTVFPPTSFGPLVRKIFVDGVPFVVSTGYHF
||||||||||||||||||||||||| |||||||||||||||||||||||||
Mutated CVNCLDDRCILHCANFNVLFSTVFPLTSFGPLVRKIFVDGVPFVVSTGYHF
| Allele | Reference peptide | Mutated peptide | Reference affinity (IC50, nM) | Mutated affinity (IC50, nM) |
|---|---|---|---|---|
| HLA-A*02:01 | FPPTSFGPLV | FPLTSFGPLV | 8999 | 21 |
| HLA-A*02:02 | FPPTSFGPLV | FPLTSFGPLV | 7237 | 18 |
| HLA-A*02:11 | FPPTSFGPLV | FPLTSFGPLV | 3405 | 9 |
| HLA-A*32:01 | VLFSTVFPP | VLFSTVFPL | 5415 | 27 |
| HLA-A*02:11 | LFSTVFPP | LFSTVFPL | 6706 | 35 |
| HLA-A*02:11 | ANFNVLFSTVFPP | ANFNVLFSTVFPL | 1827 | 12 |
| HLA-A*02:11 | NFNVLFSTVFPP | NFNVLFSTVFPL | 1779 | 13 |
| HLA-A*02:11 | NVLFSTVFPP | NVLFSTVFPL | 739 | 6 |
| HLA-C*03:03 | VLFSTVFPP | VLFSTVFPL | 28468 | 265 |
| HLA-C*03:04 | VLFSTVFPP | VLFSTVFPL | 28468 | 265 |
| HLA-A*02:11 | FNVLFSTVFPP | FNVLFSTVFPL | 413 | 4 |
| HLA-C*14:02 | VLFSTVFPP | VLFSTVFPL | 21407 | 220 |
| HLA-A*02:11 | FNVLFSTVFPPT | FNVLFSTVFPLT | 2618 | 27 |
| HLA-A*02:01 | LFSTVFPP | LFSTVFPL | 7475 | 81 |
| HLA-C*14:02 | LFSTVFPP | LFSTVFPL | 11979 | 138 |
| HLA-B*15:03 | VLFSTVFPP | VLFSTVFPL | 3115 | 36 |
| HLA-A*02:11 | VLFSTVFPP | VLFSTVFPL | 78 | 1 |
| HLA-C*03:02 | VLFSTVFPP | VLFSTVFPL | 20533 | 273 |
| HLA-A*02:01 | FNVLFSTVFPP | FNVLFSTVFPL | 656 | 9 |
| HLA-A*02:01 | ANFNVLFSTVFPP | ANFNVLFSTVFPL | 2569 | 37 |
| HLA-A*02:11 | FNVLFSTVFPPTS | FNVLFSTVFPLTS | 9448 | 142 |
| HLA-A*02:02 | NVLFSTVFPP | NVLFSTVFPL | 1088 | 17 |
| HLA-A*02:11 | CANFNVLFSTVFPP | CANFNVLFSTVFPL | 6143 | 96 |
| HLA-A*02:02 | FNVLFSTVFPP | FNVLFSTVFPL | 1014 | 16 |
| HLA-A*68:02 | NVLFSTVFPP | NVLFSTVFPL | 2213 | 37 |
| HLA-A*02:02 | ANFNVLFSTVFPP | ANFNVLFSTVFPL | 3126 | 53 |
| HLA-A*02:11 | PPTSFGPLV | PLTSFGPLV | 28329 | 483 |
| HLA-C*16:01 | VLFSTVFPP | VLFSTVFPL | 24733 | 432 |
| HLA-C*17:01 | VLFSTVFPP | VLFSTVFPL | 25087 | 439 |
| HLA-A*02:01 | FNVLFSTVFPPT | FNVLFSTVFPLT | 3765 | 66 |
| HLA-A*02:02 | LFSTVFPP | LFSTVFPL | 9311 | 167 |
| HLA-A*02:01 | NVLFSTVFPP | NVLFSTVFPL | 928 | 17 |
| HLA-A*02:02 | NFNVLFSTVFPP | NFNVLFSTVFPL | 2542 | 47 |
| HLA-A*02:02 | VLFSTVFPP | VLFSTVFPL | 269 | 5 |
| HLA-A*02:01 | NFNVLFSTVFPP | NFNVLFSTVFPL | 2286 | 44 |
| HLA-A*02:06 | FPPTSFGPLV | FPLTSFGPLV | 3379 | 73 |
| HLA-A*02:01 | VLFSTVFPP | VLFSTVFPL | 179 | 4 |
| HLA-A*02:06 | FNVLFSTVFPP | FNVLFSTVFPL | 1609 | 37 |
| HLA-A*02:02 | FNVLFSTVFPPT | FNVLFSTVFPLT | 4116 | 110 |
| HLA-A*02:11 | FSTVFPPTSFGPLV | FSTVFPLTSFGPLV | 6695 | 180 |
| HLA-A*02:01 | FNVLFSTVFPPTS | FNVLFSTVFPLTS | 11451 | 311 |
| HLA-A*02:06 | VLFSTVFPP | VLFSTVFPL | 368 | 10 |
| HLA-A*02:11 | ANFNVLFSTVFPPT | ANFNVLFSTVFPLT | 14639 | 409 |
| HLA-A*02:01 | FSTVFPPTSFGPLV | FSTVFPLTSFGPLV | 12596 | 425 |
| HLA-A*68:01 | PTSFGPLVR | LTSFGPLVR | 908 | 36 |
| HLA-A*02:06 | NVLFSTVFPP | NVLFSTVFPL | 1611 | 66 |
| HLA-B*15:01 | VLFSTVFPP | VLFSTVFPL | 5731 | 243 |
| HLA-B*35:01 | FPPTSFGPL | FPLTSFGPL | 304 | 13 |
| HLA-B*53:01 | FPPTSFGPL | FPLTSFGPL | 3581 | 154 |
| HLA-C*01:02 | FPPTSFGPL | FPLTSFGPL | 217 | 4855 |
| HLA-A*02:01 | CANFNVLFSTVFPP | CANFNVLFSTVFPL | 8519 | 384 |
| HLA-A*02:02 | FNVLFSTVFPPTS | FNVLFSTVFPLTS | 10920 | 497 |
| HLA-A*02:02 | FSTVFPPTSFGPLV | FSTVFPLTSFGPLV | 6958 | 322 |
| HLA-A*02:11 | VFPPTSFGPLV | VFPLTSFGPLV | 4242 | 199 |
| HLA-A*02:02 | CANFNVLFSTVFPP | CANFNVLFSTVFPL | 8183 | 391 |
| HLA-A*02:11 | NFNVLFSTVFPPT | NFNVLFSTVFPLT | 9111 | 472 |
| HLA-A*02:06 | ANFNVLFSTVFPP | ANFNVLFSTVFPL | 5191 | 304 |
| HLA-A*02:06 | NFNVLFSTVFPP | NFNVLFSTVFPL | 4220 | 274 |
| HLA-A*02:06 | FNVLFSTVFPPT | FNVLFSTVFPLT | 4868 | 388 |
| HLA-B*35:03 | FPPTSFGPL | FPLTSFGPL | 286 | 24 |
| HLA-A*03:01 | PTSFGPLVRK | LTSFGPLVRK | 554 | 47 |
| HLA-A*31:01 | PTSFGPLVR | LTSFGPLVR | 4402 | 390 |
| HLA-A*11:01 | PTSFGPLVR | LTSFGPLVR | 4068 | 379 |
| HLA-A*03:02 | PTSFGPLVRK | LTSFGPLVRK | 868 | 88 |
| HLA-B*07:02 | STVFPPTSFGPL | STVFPLTSFGPL | 777 | 79 |
| HLA-A*68:01 | PTSFGPLVRK | LTSFGPLVRK | 308 | 34 |
| HLA-B*07:02 | TVFPPTSFGPL | TVFPLTSFGPL | 1498 | 171 |
| HLA-A*02:11 | STVFPPTSFGPLV | STVFPLTSFGPLV | 2427 | 285 |
| HLA-A*02:11 | NVLFSTVFPPT | NVLFSTVFPLT | 1183 | 139 |
| HLA-A*33:03 | PTSFGPLVR | LTSFGPLVR | 3228 | 381 |
| HLA-A*26:01 | STVFPPTSF | STVFPLTSF | 786 | 95 |
| HLA-B*07:02 | VFPPTSFGPL | VFPLTSFGPL | 339 | 43 |
| HLA-A*02:01 | NVLFSTVFPPT | NVLFSTVFPLT | 2111 | 290 |
| HLA-B*42:01 | FSTVFPPTSFGPL | FSTVFPLTSFGPL | 1320 | 185 |
| HLA-B*42:01 | STVFPPTSFGPL | STVFPLTSFGPL | 738 | 111 |
| HLA-A*02:01 | VLFSTVFPPTSF | VLFSTVFPLTSF | 2225 | 342 |
| HLA-B*08:01 | FPPTSFGPL | FPLTSFGPL | 973 | 153 |
| HLA-B*42:01 | TVFPPTSFGPL | TVFPLTSFGPL | 843 | 142 |
| HLA-A*11:01 | PTSFGPLVRK | LTSFGPLVRK | 123 | 21 |
| HLA-B*42:01 | VFPPTSFGPL | VFPLTSFGPL | 274 | 47 |
| HLA-A*02:01 | VLFSTVFPPTS | VLFSTVFPLTS | 708 | 124 |
| HLA-B*07:02 | FSTVFPPTSFGPL | FSTVFPLTSFGPL | 2537 | 466 |
| HLA-B*07:02 | FPPTSFGPL | FPLTSFGPL | 42 | 8 |
| HLA-A*02:11 | VLFSTVFPPTSF | VLFSTVFPLTSF | 1019 | 195 |
| HLA-A*25:01 | STVFPPTSF | STVFPLTSF | 1533 | 296 |
| HLA-B*07:02 | FPPTSFGPLV | FPLTSFGPLV | 1131 | 236 |
| HLA-B*42:01 | FPPTSFGPL | FPLTSFGPL | 21 | 5 |
| HLA-A*26:01 | FSTVFPPTSF | FSTVFPLTSF | 1667 | 398 |
| HLA-B*51:01 | FPPTSFGPL | FPLTSFGPL | 1809 | 433 |
| HLA-A*02:11 | VLFSTVFPPTS | VLFSTVFPLTS | 309 | 74 |
| HLA-B*42:01 | FPPTSFGPLV | FPLTSFGPLV | 368 | 89 |
| HLA-A*02:02 | VLFSTVFPPTS | VLFSTVFPLTS | 1210 | 377 |
| HLA-C*03:02 | FPPTSFGPL | FPLTSFGPL | 960 | 313 |
| HLA-A*32:01 | STVFPPTSF | STVFPLTSF | 230 | 86 |
| HLA-B*58:01 | FSTVFPPTSF | FSTVFPLTSF | 88 | 33 |
| HLA-B*15:01 | STVFPPTSF | STVFPLTSF | 130 | 49 |
| HLA-A*02:01 | VLFSTVFPPT | VLFSTVFPLT | 61 | 23 |
| HLA-C*03:03 | FPPTSFGPL | FPLTSFGPL | 375 | 145 |
| HLA-C*03:04 | FPPTSFGPL | FPLTSFGPL | 375 | 145 |
| HLA-A*02:02 | VLFSTVFPPT | VLFSTVFPLT | 113 | 44 |
| HLA-C*16:01 | FSTVFPPTS | FSTVFPLTS | 1127 | 460 |
| HLA-C*14:02 | LFSTVFPPTSF | LFSTVFPLTSF | 396 | 950 |
| HLA-C*01:02 | VFPPTSFGPL | VFPLTSFGPL | 429 | 997 |
| HLA-A*02:02 | TVFPPTSFGPLV | TVFPLTSFGPLV | 789 | 343 |
| HLA-A*68:02 | STVFPPTSFGPLV | STVFPLTSFGPLV | 169 | 74 |
| HLA-A*02:11 | VLFSTVFPPT | VLFSTVFPLT | 25 | 11 |
| HLA-A*03:01 | TVFPPTSFGPLVRK | TVFPLTSFGPLVRK | 92 | 198 |
| HLA-C*14:02 | VFPPTSFGPL | VFPLTSFGPL | 47 | 99 |
| HLA-A*23:01 | LFSTVFPPTSF | LFSTVFPLTSF | 370 | 179 |